Abstract
Ranaviruses (RV, Iridoviridae) are now known to infect fish, amphibians, and reptiles, raising considerable ecological and commercial concerns due to the escalating infection prevalence and the resulting die-offs of wild and aquacultural species. Notably, ranaviruses exhibit uncanny capacities to cross host species barriers of their poikilothermic hosts, likely owing to their potent immune evasion mechanisms. In turn, the species infected by these pathogens possess immune systems that are less well understood than those of mammals, and are often comprised of unique immune genes or multiple copy orthologs of the single hallmark mammalian immune factors. Thus, garnering greater insight into ranavirus infection strategies is largely contingent on gaining further insights into host immune barriers faced by these emerging infectious agents. Accordingly, here we coalesce the current state of understanding of the distinct facets of lower vertebrate immune responses to ranaviral infections and underline some of the evasion strategies by which these pathogens circumvent these host defenses.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
- Viral Hemorrhagic Septicemia Virus
- Infectious Pancreatic Necrosis Virus
- Adult Frog
- Antiviral Immunity
- Antiviral Immune Response
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Infections of ectothermic vertebrates by members of the genus Ranavirus (RV; family Iridoviridae) and the resulting disease outbreaks and die-offs among wild and farmed populations have escalated at alarming rates recently and raised considerable concerns. While it is apparent that individual teleost, amphibian, and reptile species vary in their susceptibility to these pathogens, the immune and viral determinants of ranaviral diseases are at present unclear. In fact, with the rapid rise in both the prevalence of ranavirus infections and the remarkable capacity of these viruses to infect new hosts, ranaviruses such as Frog Virus 3 (FV3) are now considered to be a potential global threat to poikilothermic populations (Gray and Miller 2013). There is a pressing need to determine whether the susceptibility of a given ectothermic species reflects its inability to mount a protective antiviral immune response or the capacity of the ranavirus to overcome otherwise intact immune barriers. Indeed, ranaviruses appear to possess an array of immune evasion and host modulation mechanisms. Thus, a more thorough examination of the ranavirus-host immune interface at the molecular and cellular levels is necessary in order to devise potential preventative measures against these viral agents.
Compared to mammals, ectothermic vertebrates possess a complex immune system, but they mount relatively less effective adaptive immune responses. Ectotherms display poorer T lymphocyte expansion, fewer antibody isoforms, and generally a less developed immunological memory response than mammals (Robert and Ohta 2009). Thus, these organisms may rely more heavily on innate immune responses to facilitate clearance of pathogens such as ranaviruses. Innate antiviral immune defenses in ectothermic vertebrates are distinct from those described in mammals, although the exact contribution and efficacies of this immune response need to be elucidated in the context of ranavirus infections.
This chapter summarizes recent advances in our understanding of the contributions of innate and adaptive immune responses to the elimination and/or progression of ranaviral infections in poikilotherms as well as an overview of the strategies of these pathogens for evading host immune barriers.
2 Innate Immune Responses to Ranavirus Infections
2.1 Antimicrobial Peptide Responses to Ranaviral Infection
Antimicrobial peptides are an important element of anuran innate immunity that provides protection to skin and mucosal surfaces against a variety of pathogens. These small molecules are synthesized and stored in the dermal granular glands and secreted into mucus in response to stress or injury (Rollins-Smith 2009; Rollins-Smith et al. 2005). Antimicrobial peptides are also involved in defenses against ranaviruses. Esculentin-2P (E2P) and Ranatuerin-2P (R2P), two antimicrobial peptides isolated from Rana pipiens, are capable of inactivating both FV3 and channel catfish virus (CCV) within minutes and at temperatures as low as 0 °C. This suggests that direct interaction of these molecules with the viruses rather than inhibition of viral replication is responsible for the drop in infectivity (Chinchar et al. 2001). The ability of antimicrobial peptides to function across a broad range of temperatures presumably reflects the ectothermic nature of the host. Notably, 50 μM of E2P or R2P was sufficient for 99% inactivation of CCV, whereas a ten times greater concentration of either peptide was necessary to achieve 90% inactivation of FV3 (Chinchar et al. 2001). It was postulated that the greater resistance of FV3 to inactivation reflected the difficulty of antimicrobial peptides to target the inner lipid membrane beneath the FV3 capsid. Presumably, this inner membrane requires disruption in order for viral inactivation to occur. Other antimicrobial peptides, including Ranatuerin-2YJ, Dybowskin-YJb, Dybowskin-YJa, Temperin-YJa, and Temperin-YJb have been identified and cloned from the skin of Rana dybowskii infected with Rana grylio virus (RGV) (Yang et al. 2012). Interestingly, all of these peptides conferred concentration-dependent inhibition of RGV plaque formation, while viral clearance coincided with increased expression of genes encoding these molecules (Yang et al. 2012).
2.2 Innate and Inflammatory Immune Responses
Ranaviral infections are widely associated with prominent host inflammatory responses. Indeed, akin to mammalian viral infections, ranavirus-elicited inflammatory responses represent a double-edged sword as they are both critical for viral clearance, but may also exacerbate ranavirus-mediated disease and adversely affect host survival. As it stands, there is substantial documentation of innate immune responses and associated inflammation to ranavirus infections across a range of poikilothermic host species (Carey et al. 1999; Chen and Robert 2011; Grayfer et al. 2014; Jancovich and Jacobs 2011; Morales et al. 2010).
2.2.1 Anuran Amphibians
Our research group has adopted and optimized the infection of Xenopus laevis by FV3 as a model for ranavirus-ectothermic vertebrate (particularly anuran) antiviral immunity. This model pairs FV3, the best-described ranavirus at the molecular level, with Xenopus laevis, which possesses the most-characterized amphibian immune system. Typically, our experimental approach involves the intraperitoneal (i.p.) injection of X. laevis adults or tadpoles with FV3 followed by an assessment of the progress of infection, viral replication, and the host immune response. Although similar immune responses may be obtained in Xenopus by using i.p. injection or water bath exposure (Robert et al. 2011), the former tends to be more convenient and consistent for immunological studies. By this approach, we have been able to delineate the sequential progression of the innate and adaptive immune responses of adult X. laevis throughout the course of FV3 infection (Morales et al. 2010). In X. laevis adults, histochemical and flow cytometric analyses revealed that activated mononuclear and polymorphonuclear phagocytes are recruited to, and heavily represented within peritoneal exudates as early as 1 day following i.p. infection (Morales et al. 2010). We subsequently observed the peritoneal recruitment and accumulation of natural killer (NK) cells by 3 days after infection, whereas lymphocyte recruitment, including the increased presence of T cells, was not observed until 6 days post-FV3 challenge (Morales et al. 2010). Notably, the rapid accumulation of peritoneal leukocytes was concomitant with substantially elevated inflammatory gene expression. Among the hallmark inflammatory genes examined, we observed significant increases in the expression of X. laevis tumor necrosis factor-alpha (TNF-α) as early as 1 day postinfection and persisting to 3 days after FV3 exposure (Morales et al. 2010). Expression of the interleukin-1-beta (IL-1β) gene, encoding an early proinflammatory cytokine produced by macrophages, was elevated at days 1 through 6 of FV3 challenge, while the anti-inflammatory arginase-1 (Arg-1), a marker of alternatively polarized (M2) macrophages (Joerink et al. 2006b, c), was elevated at day 1 post-viral challenge and subsequently decreased (Morales et al. 2010). Together these findings show an effective and well-coordinated antiviral immune response, with sequential recruitment of innate and adaptive immune cell effectors and corresponding immune gene activation. The elevated level of Arg-1 gene expression at 1 day postinfection may be reflective of resident, rather than recruited inflammatory myeloid populations. Indeed, subsequent to FV3 peritoneal inoculation, we have consistently observed elevated mRNA transcripts for macrophage and granulocyte colony-stimulating factor receptors (M-CSFR and G-CSFR, respectively), indicative of accumulating myeloid infiltrates (L. Grayfer and J. Robert, University of Rochester, unpublished data). Notably, the elevated expression of M-CSFR (and G-CSFR) within peritoneal leukocytes (PLs) is typically accompanied by significantly increased expression of the M1 macrophage marker, inducible nitric oxide synthase (iNOS), which catalyzes the production of the antimicrobial nitric oxide by inflammatory macrophages (L. Grayfer, F. De Jesús Andino and J. Robert, University of Rochester, unpublished data). This supports the observation of decreased Arg-1 expression with the onset of an inflammatory state within the peritoneum and indicates that Arg-1 and iNOS functions are opposite across multiple groups of vertebrates (Joerink et al. 2006a, b, c; Wiegertjes and Forlenza 2010).
It is important to emphasize that amphibian susceptibility to ranaviruses varies considerably among species, their respective stages of development, and even between different populations of the same species (Miller et al. 2011). These differences likely result from multiple complex determinants including host and ranavirus genetic variability as well as the respective host immune status. Extensive immunological studies of Xenopus suggest that tadpoles possess a distinct immune system from that of adults. The larval system is typically more immature, particularly with regard to adaptive immunity (e.g., poor T cell and antibody responses). In this regard, it is altogether not surprising that tadpoles are typically unable to fully control ranavirus infections and succumb to these pathogens (Bayley et al. 2013; Grayfer et al. 2014; Hoverman et al. 2010; Landsberg et al. 2013; Reeve et al. 2013). However, it is clear that in some amphibian species, adult frogs also die from ranavirus infections (Sutton et al. 2014). Indeed, several reports indicate that compared to larvae of given amphibian species, metamorphic (Brunner et al. 2004; Haislip et al. 2011; Reeve et al. 2013) and adult (Duffus et al. 2013) animals may be more susceptible to ranaviruses. Given the rapid development of tadpoles and their drastic metamorphic remodeling, species-specific immune development pathways may explain differences in ranavirus susceptibility. Extensive immune remodeling may render metamorphs more susceptible than larvae at critical developmental stages. Moreover, the fact that ranavirus-infected Xenopus tadpoles bear lower viral loads than adults, yet succumb more readily to FV3 infection (Grayfer et al. 2014) suggests that it may be more difficult to detect this virus in tadpoles than adults or that lower viral loads trigger markedly greater disease in tadpoles.
To delineate possible inefficiencies in innate immune responses of anuran tadpoles that may account for ranavirus susceptibility, we performed an extensive comparison of immune gene expression patterns between FV3-infected X. laevis tadpoles and adults (De Jesús Andino et al. 2012). In contrast to infected adult frogs, tadpoles exhibited poor and considerably delayed anti-FV3 inflammatory gene responses (De Jesús Andino et al. 2012). TNFα, IL-1β, and IFNγ gene expression in tadpole PLs, splenocytes, and kidneys did not significantly increase until 6 days post FV3 challenge, which is in contrast with the robust and quick (1 dpi) upregulation of these genes in infected adults (De Jesús Andino et al. 2012). Notably, stimulation of tadpoles with heat-killed Escherichia coli readily elicits rapid induction of the above genes within 24 h, suggesting that the immune delays are FV3 specific (De Jesús Andino et al. 2012). These inefficiencies in the tadpole innate immune response to FV3 may reflect multiple nonexclusive issues including viral immune evasion, defect(s) in the tadpole pathogen sentinel receptor system, or physiological treadoffs to forego energetically costly inflammatory responses in favor of growth and development. Thus, these modest and delayed immune responses are likely contributing factors for the characteristically higher susceptibility of anuran tadpoles to FV3 infection and severe disease.
2.2.2 Urodel Amphibians
Consistent with the notion that hosts mount broad inflammatory responses to ranaviruses, a comprehensive microarray analysis of axolotls (Ambystoma mexicanum) infected with the Ambystoma tigrinum virus (ATV) revealed the upregulation of numerous hallmark pro-inflammatory and innate immune gene components in the spleens and lungs of these animals (Cotter et al. 2008). These genes included (but were not limited to) phagocytic receptors and intracellular components, cytokine signaling molecules, complement components, NADPH oxidase subunits (myeloid enzyme catalyzing the reactive oxygen antimicrobial response), and myloperoxidase (granulocyte enzyme catalyzing the production of hydrogen peroxide) (Cotter et al. 2008). In contrast to what has been observed in X. laevis infected with FV3 (Morales et al. 2010; Morales and Robert 2007), no lymphocyte proliferation genes were upregulated in response to ATV infections (Cotter et al. 2008). This lack of an efficient adaptive response in this species may explain why ATV is so lethal to urodels. Alternatively these observations may reflect a different infection strategy by ATV.
2.2.3 Teleost Fish
There is a substantial literature documenting innate immune and associated inflammatory responses to ranavirus infections in bony fish. Infection of the Epithilioma papulosum cyprini (EPC) teleost cell line with four distinct ranaviruses, FV3, European catfish virus (ECV), Doctor fish virus (DFV), and Epizootic haematopoietic necrosis virus (EHNV), resulted in distinct inflammatory gene expression profiles (Holopainen et al. 2012). Specifically, EHNV and FV3 elicited expression of the hallmark pro-inflammatory genes, TNFα and IL-1β, whereas ECV and DFV induced the transient expression of a generally immunosuppressive gene, transforming growth factor-beta (TGFβ) (Holopainen et al. 2012). Interestingly, all four viruses elicited expression of apoptotic components and β2-microglobulin, which is critical for surface MHC class I expression and cytotoxic T cell function, suggesting that at least with respect to FV3 infection of teleosts (as compared to axolotls), the adaptive immune response may be elicited by ranaviral infections.
A recent microarray study examined the transcriptional response of fathead minnow (FHM) cells following infection with either wild type (wt) FV3 or a knockout (KO) mutant lacking the truncated vIF-2α gene. Infection with wt FV3 resulted in the upregulation of numerous immune related genes by 8 h p.i., including IL-8, IFN, IFN regulatory factor (IRF) -1, -2, and -3, IL-1β, etc. For the most part, similar genes were upregulated in cells infected with the KO mutant, but the magnitude of the induction was generally lower (Cheng et al. 2014).
2.3 Determinants of Ranavirus-Induced Pathogenicity and Mortality
An appropriate, timely resolution of inflammation is just as important as the induction and progression of this response, because a prolonged inflammatory response increases the risk of tissue damage and host death (Fullerton et al. 2013). Although sparse, there is evidence suggesting that ranavirus infections may exacerbate inflammatory responses, accounting for some of the observed ranavirus pathology. For example, in 1997 a novel iridovirus was isolated in Saskatchewan (Canada) from larval tiger salamanders (Bollinger et al. 1999). These animals suffered from exacerbated inflammation, necrosis, and characteristic ranavirus-induced cytoplasmic inclusions within splenic, renal, lymphoid, and hematopoietic tissues (Bollinger et al. 1999). Similarly, whole populations of ranavirus-infected green-striped tree dragons (Japalura splendida) exhibited systemic hemorrhaging, necrosis, granulomatous, and necrotic inflammation, as well as severe renal pathology, hyperanemia, and extensive hepatic damage (Behncke et al. 2013), culminating in mass mortality. Ranavirus infection within pythons suggests that inflammation may be a determinant of ranaviral pathology (Hyatt et al. 2002). Mortality of largemouth bass infected intraperitoneally with largemouth bass virus (LMBV) is believed to result from virally induced inflammation and associated necrosis (Zilberg et al. 2000). Consistent with these inflammatory symptoms, juvenile bass inoculated with LMBV exhibited corkscrew swimming and distended abdomens (Zilberg et al. 2000). Notably, the deeper tissues of infected fish were unaffected, bringing into question whether virus-induced damage was due to target cell accessibility or the limitations of LMBV cell tropism. The latter suggests that inflammation and necrotic damage resulting in mortality may be the result of primary injuries at the initial sites of infection. Indeed, the above observations are reminiscent of earlier studies of FV3 infections in rodents (Gut et al. 1981; Kirn et al. 1980, 1982), in which, despite the inability of FV3 to replicate at 37 °C (Aubertin et al. 1973), the initial viral inoculum was responsible for extensive inflammation, necrosis, and liver damage.
Our recent findings in Xenopus support the induction of a pro-inflammatory response by FV3 (Grayfer et al. 2014). Mindful of the idea that X. laevis adults presumably mount effective anti-ranaviral responses leading to viral clearance, we were intrigued to find that (at least during acute infections) adults possessed significantly greater FV3 loads (1 to 2 orders of magnitude) than tadpoles, which are typically more susceptible to FV3 infections (Grayfer et al. 2014). Also, a possible influence of temperature on viral loads was observed in wood frog tadpoles, where these animals succumbed to infection quicker at 25 °C but at higher viral loads than those maintained at 15 °C (J. Chaney and M. Gray, University of Tennessee, unpublished data). These results suggest that FV3 virulence is not strictly dependent on the magnitude of viral replication. Additionally, immunocompetent tadpoles may be more vulnerable to ranaviral virulence factors and other environmental parameters than adults. In support of this hypothesis, we observed that although tadpoles pre-stimulated with recombinant X. laevis type I interferon (rXlIFN) possessed viral loads several logs lower than adults, they nonetheless succumbed to FV3 infection (Grayfer et al. 2014). Furthermore, despite lower FV3 loads, IFN-treated larvae experienced damage to multiple organs, including extensive loss of tissue architecture and cellular organization through necrosis and apoptosis, albeit without extensive leukocyte infiltration (Grayfer et al. 2014). Therefore, even at markedly reduced viral loads, ranaviruses may confer irreversible tissue damage in tadpoles relatively early in infection, resulting primarily from virus-mediated cytopathology rather than from viral replication. Indeed, as seen in rodent models of FV3, ranaviruses may trigger toxic and potentially lethal effects, irrespective of their capacity to replicate within their host cells (Gendrault et al. 1981). This notion has recently been substantiated with Grouper iridovirus (GIV). Replication-deficient UV-inactivated GIV induced apoptosis in two of the three infected cell lines (Pham et al. 2012). Similarly, heat- and UV-inactivated FV3 elicits FHM cell apoptosis and inhibits host RNA and protein synthesis (Chinchar et al. 2003; Raghow and Granoff 1979). Based on these findings, we hypothesize that inoculation of animals with sufficient inactivated virus will induce toxicity in the absence of virus replication. If this hypothesis holds true for other members of the genus and family, we may need to consider that these viruses are more pathogenic than previously thought.
3 The Complex Roles of Macrophage-Lineage Cells in Ranaviral Disease
3.1 Inferences from Rodent Models of FV3 Infection
The involvement of macrophage-lineage cells in ranavirus infections may be inferred from initial studies conducted 30 years ago using rodents as models of hepatitis (Gut et al. 1981; Kirn et al. 1980, 1982). These early studies revealed that Kupffer cells (liver macrophages) were the principal targets of FV3 infection and that their death was linked to the loss of hepatic clearance and culminated in severe hepatitis and mortality (Gut et al. 1981). These studies also implicated inflammation as a contributor to FV3-mediated pathology, including extensive leukotriene release by Kupffer cells (Hagmann et al. 1987). Inhibition of leukotriene synthesis within FV3-infected animals dramatically reduced virally elicited hepatic damage (Hagmann et al. 1987), suggesting that pathology was largely due to inflammatory responses.
Although FV3 is not a mammalian pathogen and, with the exception of expression of select early genes (Lopez et al. 1986) does not replicate at 37 °C (Aubertin et al. 1973), this work nonetheless supports the current hypothesis that, because of their high phagocytic and endocytic activity, macrophage-lineage cells are integral targets for ranavirus infections. In fact, the absence of replication at 37 °C may be viewed as an advantage for investigating the mechanisms of FV3 cell entry. In cultured rat Kupffer cells, viral particles appeared in phagocytic vacuoles and endocytic compartments promptly following FV3 infection (Gendrault et al. 1981). Moreover, a substantial proportion of FV3 virions that attached to cells displayed viral capsid-host membrane fusion and release of viral core contents into cell cytoplasm (Gendrault et al. 1981). This observation suggests that the underlying mechanisms governing ranavirus entry are universal and facilitate entry of cells from organisms as evolutionarily distant as mammals, fish, and amphibians. In line with this reasoning, it is likely that cells of the myeloid lineage serve as ranaviral targets precisely because of their high efficiency of ingestion of extracellular materials, facilitated by an array of endocytic/phagocytic surface receptors, several of which likely recognize and bind ranaviruses. This feature of vertebrate professional phagocytes may have been targeted as a ranaviral infection strategy and may explain why ranaviruses successfully cross host species boundaries. Furthermore, because ranaviruses cannot replicate at mammalian body temperatures, the above-described literature implies that that the pathological events seen in FV3-infected rodents are not the result of full virus replication. Instead, cell death is presumably triggered by preformed lytic factors encapsulated within FV3 virions or the expression of early viral gene products (Lopez et al. 1986). Similar to the mRNA present within adenovirus virions (Chung et al. 2003), FV3 early gene expression at nonpermissive temperatures may also result from the release of prepackaged virulence factor-encoding mRNAs rather than from de novo viral transcription. Indeed, FV3 infection of mammalian cells induces rapid cellular RNA, DNA, and protein synthesis arrest (Elharrar et al. 1973). Furthermore, factors solubilized from FV3 virions result in cellular toxicity and inhibit host macromolecular synthesis (Aubertin et al. 1973; Kirn et al. 1972)
3.2 Amphibian Vectors of Ranaviral Dissemination and Persistence
Increasing evidence from natural ranavirus infections of amphibians supports the idea that macrophages are important not only for antiviral defense, but also to ranavirus infection strategies. We utilized FV3 infection of X. laevis as a platform for the study of the ranavirus-host immune interface, with converging lines of evidence from our past and current work confirming FV3-X. laevis macrophage interactions. We have demonstrated that FV3 persists within amphibian hosts for several months following the resolution of clinically apparent disease (Robert et al. 2007). In addition, FV3 DNA could be detected in healthy animals that were not infected in the laboratory. This suggests that FV3 adopts some form of quiescence as a means of maintenance within immuno-competent hosts. Notably, FV3 effectively infects frog peritoneal leukocytes both in vitro and in vivo, persists within these cells and undergoes active viral transcription for up to 12 days subsequent to infection (Robert et al. 2007). Since peritoneal leukocytes are comprised predominantly of macrophage-lineage cells, our findings not only corroborate the ranavirus-macrophage tropism, but also suggest that these terminally differentiated, long-lived populations are ideal vectors for viral dissemination, or serve as “within host” reservoirs.
The above hypothesis has been substantiated by our subsequent transmission electron microscopy analysis of FV3-infected X. laevis peritoneal leukocytes in which we detected icosahedral virus particles in peritoneal leukocytes bearing macrophage morphology (Morales et al. 2010). These FV3-infected cells exhibited small numbers of intracellular viral particles, implying that FV3 may employ mononuclear phagocytes as a reservoir for dissemination. FV3-macrophage interaction is reminiscent of the HIV-macrophage relationship in which viral particles accumulate within the myeloid cells as a mechanism of dissemination (Coiras et al. 2009; Goodenow et al. 2003; Gousset et al. 2008; Groot et al. 2008). Interestingly, several of these FV3-infected peritoneal macrophages not only contained cytoplasmic virions, they also shed virions into the extracellular milieu (Morales et al. 2010), confirming that these cells likely function as both reservoirs and vectors of viral dissemination within their hosts.
In general, i.p. infection of adult X. laevis with FV3 leads to extensive recruitment of leukocytes, including a large number of cells with macrophage morphology, to the site of infection (Morales et al. 2010). The myeloid origins of these leukocytes are supported not only by their expression of macrophage inflammatory genes (TNFα, IL-1β, and Arginase-1; Morales et al. 2010), but also of the macrophage-lineage marker, M-CSFR (L. Grayfer, F. De Jesús Andino and J. Robert, University of Rochester, unpublished data). Interestingly, while we have been able to amplify FV3 DNA from peritoneal leukocytes isolated from virus-infected frogs up to 21 days postinfection, FV3 early and late transcripts were detected at 6, but not 15 or 21 days postinoculation (Morales et al. 2010). This suggests that the viral genome is maintained within macrophage-like cells in a state of dormancy. Possibly, the inability to detect FV3 genomes among peritoneal leukocytes at later times may reflect the dissemination of these cells to distal sites within the X. laevis host.
In Xenopus, the kidney represents a focal site of FV3 replication. Interestingly, active FV3 gene transcription is seen in some, but not all FV3-infected frogs for up to 9 days after infection, whereas viral genomic DNA may be amplified from some of these animals 2 weeks subsequent to viral challenge (Morales et al. 2010). Notably, this interval is shorter than the 3-week period during which viral persistence is reliably detected within peritoneal leukocyte populations. These differences possibly reflect the distinct interactions between FV3 and these various cell targets. Presumably, kidney cells are productively infected and serve as sites of active FV3 replication, indicated by high viral titers and extensive tissue damage (Grayfer et al. 2014). Conversely, macrophage-lineage cells are terminally differentiated, nondividing, long-lived cells, which ostensibly serve as reservoirs for dormant (non-replicating) ranavirus.
Collectively, these observations implicate frog macrophage-lineage cells as central targets of FV3 infection, serving as likely cellular targets for persistence, quiescence, and dissemination. Indeed, we have recently observed that FV3 genomic DNA may be amplified from in vitro-infected, cultured peritoneal phagocytes without detectable viral gene expression as long as several months after initial infection (L. Grayfer, and J. Robert, University of Rochester, unpublished data). We believe that the key to further delineating ranavirus-macrophage interactions and ranaviral quiescence is contingent on developing in vitro myeloid cell cultures and related reagents.
3.3 Macrophage Reservoirs and Ranavirus Reactivation
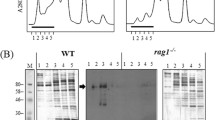
We recently provided substantial support for the hypothesis that amphibian macrophages serve not only as vehicles of ranavirus dissemination within the host but also as foci of disease reactivation. When peritoneal phagocytes were isolated from 15 X. laevis adults 30 days postinfection, only cells from one individual displayed detectable levels of FV3 DNA and expressed transcripts encoding the viral DNA polymerase and major capsid protein (J. Robert, L. Grayfer, and F. De Jesús Andino, University of Rochester, unpublished data). However, after i.p. injection of heat-killed E. coli, nine of the same 15 animals exhibited both detectable viral genomic DNA and active viral gene expression. In addition, immunofluorescence microscopy targeting 53R, an FV3 gene product required for replication and assembly, and HAM56, a X. laevis macrophage marker (Nishikawa et al. 1998), revealed that peritoneal macrophages displayed productive FV3 replication (Fig. 1). Thus, it appears that mononuclear phagocytes harbor low levels of viral DNA, which can be reactivated by inflammation. Further research into activation states (both classical and alternative; Auffray et al. 2007; Nahrendorf et al. 2007; Zhao et al. 2009; Ziegler-Heitbrock 2007) of mononuclear phagocytes will be critical to devising preventative measures and understanding the precise infection strategies of these complicated pathogens.
Xenopus laevis HAM 56+ peritoneal macrophages infected with Frog Virus 3. Xenopus laevis peritoneal leukocytes were infected at a multiplicity of infection (MOI) of 0.3 with Frog Virus 3. Macrophages were stained with an antibody against the macrophage marker HAM56. FV3 was visualized using an antibody against the 53R viral protein. Hoechst was used to visualize the cellular nuclei
3.4 Ranavirus Infections Among Other Poikilothermic Macrophages
Akin to many other pathogens, ranaviruses presumably overcome macrophage antimicrobial and antiviral barriers, at which point these cells become vehicles for both viral dissemination and persistence. However, exploitation of macrophage-lineage cells as vectors of viral dissemination and persistence does not appear confined to FV3, as other members of the genus Ranavirus and family Iridoviridae have also adopted this mechanism of host infiltration and immune evasion. For example, an iridovirus-like pathogen infects sheatfish kidney macrophages and is capable of down-regulating phorbol myristate acetate-elicited reactive oxygen production by these cells in vitro (Siwicki et al. 1999). Likewise, following infection with the Taiwan Grouper Iridovirus (TGIV), elevated numbers of phosphatase-positive, highly phagocytic basophilic and eosinophilic mononuclear leukocytes were detected (Chao et al. 2004). Interestingly, TGIV genomic DNA was found only within the nuclei of mononuclear phagocytes at early times after infection, whereas at later times it was seen in both nuclear and cytosolic compartments and these cells lost their phagocytic capacity (Chao et al. 2004). Clearly, TGIV has evolved intricate and temporally regulated strategies for overcoming and utilizing the very immune cells that would presumably be coordinating the antiviral immune response. It is probable that the strategy of invading mononuclear phagocytes as a means of immune evasion and dissemination is a distinct feature of all vertebrate iridoviruses. Further development of in vitro primary macrophage cultures derived from relevant host species and infection models will provide additional insight into these infection strategies.
4 Antiviral Immune Responses to Ranavirus Infections
4.1 Antiviral Interferons of Ectothermic Vertebrates
The interferon (IFN) response provides a significant contribution to antiviral immunity. IFN responses generally arise as the result of recognition of viral products through an array of host pathogen recognition receptors (PRRs), including toll-like receptors (TLRs), retinoic acid-inducible gene 1-(RIG-I)-like receptors (RLRs), and cytosolic DNA sensors (Baum and Garcia-Sastre 2010; Sadler and Williams 2008). This branch of antiviral immunity consists of three classes of cytokines, type I, II, and III IFNs (Sadler and Williams 2008). IFNγ, the only type II IFN of mammals (n.b., bony fish possess multiple type II IFNs; Grayfer et al. 2010) plays multiple immune and antiviral roles, whereas IFN-I and -III function predominantly as antiviral molecules. Mammalian IFN-I possesses broad cellular specificities, whereas IFN-III targets specific cell subsets (Levraud et al. 2007; Zou et al. 2007). Interestingly, while the distinct receptor systems utilized by IFN-I and -III dictate cell specificity, both cytokine families activate the same downstream Janus kinase (JAK) and signal transducer and activator of transcription (STAT) signaling pathways, culminating in similar antiviral outcomes (Sadler and Williams 2008) including the induction of antiviral genes such as protein kinase R (PKR) and Myxovirus resistance (Mx) molecules.
While these IFN responses are well studied among warm-blooded vertebrates, the cold-blooded hosts that are subject to ranavirus infection possess unique and much less understood IFN systems. The mammalian type I IFNs are encoded by intronless genes, comprising the multigene IFNα family (13 in humans) and a single IFNβ gene (Hervas-Stubbs et al. 2011). While reptiles and birds also express single exon-encoded type I IFNs (Robertsen 2006; Zou and Secombes 2011), lower vertebrate species including cartilaginous and bony fish as well as amphibians possess type I IFNs encoded by five exon/four intron transcripts and displaying marked sequence divergence from their mammalian counterparts (Chang et al. 2009; Qi et al. 2010; Robertsen 2006; Zou and Secombes 2011; Zou et al. 2007).
Presently, only the type I IFN systems of bony fish have been explored in detail. These IFNs are subdivided into two groups (group I: 2C; group II: 4C) based on cysteine patterns (Sun et al. 2009; Zou et al. 2007), and further classified into four groups (IFNa–d) according to phylogeny (Chang et al. 2009; Sun et al. 2009). Importantly, while multiple distinct mammalian IFNs confer their biological roles through the same receptor complex (Li et al. 2008; Samuel 2001), fish group I and II IFNs signal through unique receptor complexes (Aggad et al. 2009). Functional studies have been performed predominantly on group I type I fish IFNs (Aggad et al. 2009; Altmann et al. 2003; Long et al. 2004; Lopez-Munoz et al. 2009; Robertsen et al. 2003; Zou et al. 2007), and it has been demonstrated that these IFNs differ in their capacities to establish cellular antiviral states (Aggad et al. 2009; Levraud et al. 2007; Li et al. 2010; Lopez-Munoz et al. 2009). For example, salmonid IFNs a–d possess different transcriptional regulation patterns and distinct antiviral functions, as some of these cytokines are capable of eliciting potent antiviral responses while others are believed not to have antiviral functions at all (Svingerud et al. 2012). The type II IFN systems of amphibians and reptiles remain largely uncharacterized, whereas those of bony fish appear to be much more complex than that of mammals (Zou and Secombes 2011), and will not be addressed further here.
Mammalian IFN-III is comprised of interferon lambda (IFNλ) -1, -2, and -3 (also designated as IL-28A, IL-28B, and IL-29). These molecules are encoded by five exon/four intron gene transcripts and signal through a receptor system composed of the interferon lambda receptor-1 (IFNλR1) and interleukin-10 receptor-2 (IL-10R2; reviewed in reference Kotenko 2011). Intriguingly, while bona fide type III IFNs either do not exist, or have not yet been identified in bony fish, amphibians are now known to possess both type I IFNs with the same five exon/four intron gene organization as their fish counterparts, as well as true type III IFNs (Qi et al. 2010). This is especially relevant when considering that amphibians are key evolutionary intermediates between fish and mammals and inhabit both aquatic and terrestrial habitats. In fact, a hallmark characteristic of fish and amphibian type I IFNs is the five exon/four intron genomic organization, which is distinct from the reptile, avian and mammalian intronless type I IFNs (Robertsen 2006; Robertsen et al. 2003; Sun et al. 2009). Hitherto, there has been substantial debate regarding the precise phylogenetic relationship of fish IFN-I to higher vertebrate IFN-I and -III. Fish cytokines exhibit exon/intron gene organization similar to that of mammalian type III IFNs, yet possess hallmarks of higher vertebrate type I IFNs such as conserved cysteine positioning and (with the exception of the catfish IFN-I) a C-terminal CAWE motif, a conserved sequence motif found within nearly all IFNs (Lutfalla et al. 2003; Qi et al. 2010; Robertsen 2006; Zou et al. 2007). It will be interesting to determine the respective roles of these molecules in fish and amphibian antiviral immunity to RVs, particularly considering that fish appear to only have type I IFNs, while frogs possess both IFN types I and III (Qi et al. 2010).
4.2 Interferon Response to Ranavirus Infection
As described above, an important antiviral gene product synthesized during the interferon response is the Myxovirus resistance (Mx) protein (Samuel 2001). Mx proteins are believed to be pivotal to the establishment of the antiviral state conferred by IFN (Samuel 2001). Mx proteins are high molecular weight GTPases belonging to the dynamin superfamily and are known to facilitate intracellular membrane remodeling as well as intracellular trafficking (Kochs et al. 2005). As in mammals, teleost Mx proteins function as antiviral mediators, with distinct Mx isoforms from different species conferring somewhat unique antiviral effects. To date the Mx of most, but not all, fish species have proven ineffective in preventing infection by various members of the family Iridoviridae. For example, Japanese flounder Mx is capable of inhibiting the replication of two species of rhabdovirus, but is incapable of inhibiting replication of Red seabream iridovirus (RSIV, genus Megalocytivirus; family Iridoviridae) (Caipang et al. 2003). Similarly, Barramundi Mx inhibits replication of the nodavirus viral nervous necrosis virus (VNNV) and of Infectious pancreatic necrosis virus (IPNV) but fails to show antiviral effects against Taiwan grouper iridovirus (TGIV) (Wu et al. 2012; Wu and Chi 2007). Likewise, Senegalese sole Mx confers antiviral effects against the IPNV and Viral hemorrhagic septicemia virus (VHSV, family Rhabdoviridae), but not against the ranavirus European sheatfish virus (ESV) (Alvarez-Torres et al. 2013). Finally, rainbow trout Mx1 is antiviral towards IPNV, Salmonid alpha virus (SAV, Togaviridae), and infectious hematopoetic necrosis virus (IHNV, Rhabdoviridae), but is not effective at blocking replication of EHNV (Lester et al. 2012; Trobridge et al. 1997). Possibly, the host antiviral responses coevolved with local ranaviral isolates. Thus, the inadequacy of antiviral components such as Mx1 in dealing with foreign ranaviral isolates may culminate in a global threat represented by geographically distant ranavirus strains introduced by subclinically infected migratory hosts or imported due to international trade.
Perhaps the most commercially and aquaculturally important fish species in southern Europe is the gilthead seabream, at least in part because of its natural resistance to most viral pathogens (Cano et al. 2006, 2009). In fact, the only viral disease affecting commercial seabream populations is Lymphocystis disease virus (LCDV, genus Lymphocystivirus, family Iridoviridae) (Cano et al. 2013). Interestingly, seabream possess at least three Mx proteins. One Mx isoform effectively inhibits replication of VHSV and LCDV, a second Mx molecule effectively inhibits replication of European sheatfish virus and LCDV, and the third is protective against VHSV (Alvarez-Torres et al. 2013; Fernandez-Trujillo et al. 2013). This represents the first example of a teleost Mx molecule effectively inhibiting DNA virus infection. This is interesting, considering that LCDV nonetheless plagues this species. It is noteworthy that in contrast to the mortality caused by many members of the family Iridoviridae, seabream effectively clear LCDV infections, although it is believed that they may harbor the virus non-symptomatically. Thus, the efficacy of the teleost IFN/Mx response may well dictate the susceptibility of individual fish species to highly virulent pathogens such as iridoviruses. Notably, many fish species are infected by, and clear LCDV. Since these infections involve fish skin (Leibovitz 1980), systemic antiviral responses such as Mx may be less important to the resolution of LCDV.
In another example, Japanese flounder IFN-inducible transmembrane (IFITM) protein is upregulated in response to Rana grylio virus (RGV) infections (Zhu et al. 2013). Furthermore, through overexpression and siRNA knockdown studies, flounder IFITM1 was shown to play an important role in the cellular antiviral response to RGV (Zhu et al. 2013). IFITM1 functions by suppressing viral-host cell entry and targeting the Golgi apparatus (Zhu et al. 2013).
As more research is conducted on these individual antiviral components, it is becoming more apparent that there are key, previously unknown factors that participate in the antiviral response of not just poikilotherms, but all vertebrate species. We propose that these individual IFN-elicited antiviral components are most likely interdependent on entire networks of other IFN-regulated molecules. Thus, the relative potency of the antiviral IFN response, at both cellular and whole organism levels, relies on the balance of numerous cellular and molecular components. Since different fish and amphibian species are now known to possess very distinct repertoires of antiviral effector molecules, it is not surprising that these disparate organisms display very different susceptibilities to similar pathogens.
Microarray analysis of axolotls infected with ATV revealed that in addition to a multifaceted inflammatory gene response, these animals also upregulate expression of multiple antiviral interferon responsive genes (Cotter et al. 2008). Among the numerous genes elicited by ranavirus infection were Mx1 genes, antiviral helicases, interferon regulatory factors, an IFITM, and a ribonuclease (Cotter et al. 2008). However, the genes encoding axolotl type I and type III IFN remain to be identified. It will be important to delineate the precise repertoire(s) of antiviral IFNs present within the axolotl genome and examine the transcriptional regulation, as well as functional roles of these moieties during immune responses against ranaviruses such as ATV.
As described above, frogs are now known to possess both type I and type III IFN genes that are transcriptionally upregulated following virus infections (Qi et al. 2010). While there have been no reported functional studies of amphibian type III IFNs, we recently identified a X. laevis type I IFN, produced it in recombinant (rXlIFN) form and characterized this molecule in the context of FV3 infections (Grayfer et al. 2014). Pretreatment of X. laevis A6 kidney-derived epithelial cell cultures with rXlIFN significantly protected these cells against the cytolytic effects of FV3 (Grayfer et al. 2014). Although control cultures were almost entirely decimated 3 days following FV3 infection and exhibited extensive viral replication, rXlIFN-treated cultures were virtually FV3-free and thriving (Grayfer et al. 2014). Furthermore, treatment of A6 cells with rXlIFN significantly upregulated the expression of Mx1 indicating that stimulation with this cytokine elicits a cellular antiviral state (Grayfer et al. 2014). Following FV3 challenge, the type I IFN response was more robust in X. laevis adults than tadpoles (Grayfer et al. 2014). Nevertheless, tadpoles injected i.p. with rXlIFN exhibited significant increases of Mx1 gene expression in the spleen and peritoneal leukocytes (Grayfer et al. 2014). Moreover, upon FV3 challenge, rXlIFN-treated tadpoles showed decreased viral replication and transcriptional activity (Grayfer et al. 2014). Therefore, in addition to delayed innate and inflammatory-associated immune gene responses, it appears that an inadequate type I IFN response also contributes to the higher susceptibility of X. laevis tadpoles to FV3.
However, adding to the complexity of the interaction between FV3 and tadpoles is the fact that although rXlIFN-treated tadpoles exhibited prolonged mean survival times following FV3 inoculation and viral loads that were several logs lower, these animal nonetheless incurred extensive organ damage and succumbed to infection (Grayfer et al. 2014). This is consistent with the notion (as described above) that depending on the species and/or developmental stage, ranaviruses may exhibit virulence factors independent of viral replication. It is intriguing that despite the antiviral potency of X. laevis IFN, FV3 ultimately results in tadpole mortality. It will be invaluable to elucidate the respective roles of amphibian type I and III IFNs, their cognate receptor systems, and the respective roles (and possibly defects) of these components during ranavirus infections of tadpole and adult frogs.
5 Adaptive Immune Responses to Ranavirus Infections
Anti-ranavirus immune responses of lower vertebrates are multifaceted, complex, and poorly understood. However, it is becoming evident that clearance of ranaviruses is heavily contingent on successful adaptive immune responses, which have been investigated to date almost exclusively in X. laevis.
5.1 Antibody Responses to Ranavirus Infection
The amphibian organization and usage of the immunoglobulin (Ig) heavy and light chain loci are reminiscent of their mammalian counterparts, including V-(D)-J rearrangements, class-switch recombination, somatic hypermutation, and affinity maturation (Du Pasquier et al. 1989, 2000; Hsu 1998). As in mammals, the Xenopus Ig class-switch from IgM to IgY (IgG analog) is thymus-dependent and requires T cell-B cell collaboration (Blomberg et al. 1980; Turner and Manning 1974). Although affinity maturation of amphibian IgY results in only a tenfold increase, as compared to the 10,000-fold increase seen with for mammalian IgGs, it has been clearly demonstrated that Xenopus humoral immunity is a significant contributing factor to anti-ranaviral immune responses, particularly of adult frogs (Maniero et al. 2006).
Following secondary FV3 infection of X. laevis adults, animals produce substantial amounts of virus-specific IgY, first detectable 1 week after infection and peaking around 3 weeks after challenge (Gantress et al. 2003). Indeed, frogs re-infected (in the absence of adjuvant) with FV3 up to 15 months post primary infection, develop anti-FV3 specific IgY antibodies in a thymus-dependent manner that are detectable from 10 days up to 8 weeks post re-immunization (Maniero et al. 2006). Notably, FV3 is effectively neutralized by exposure to this sera in vitro (Maniero et al. 2006). In addition, administration of immune sera to naturally susceptible X. laevis tadpoles immediately preceding FV3 infection confers partial, but significant passive protection against the virus (Maniero et al. 2006). Clearly, the amphibian antibody response is integral to the clearance of ranavirus infections, while the extent to which this particular immune mechanism contributes to the ultimate anti-ranaviral immunity seen in Xenopus adults remains to be determined. These results are consistent with findings with Red seabream iridovirus (RSIV, genus Megalocytivirus) in which vaccination with inactivated virions protected fish from subsequent viral challenge (Caipang et al. 2006; Nakajima and Kunita 2005; Nakajima et al. 1999)
5.2 T Cell Responses and Immunological Memory Against Ranavirus Infections
X. laevis tadpoles express suboptimal levels of MHC class Ia protein (Du Pasquier et al. 1989) and yet their splenocytes include bona fide CD8 T cells that express the pan-T Xenopus cell-surface marker CD5 (Jurgens et al. 1995) and exhibit fully rearranged TCRα/β transcripts (Horton et al. 1998). It is possible that suboptimal class Ia protein expression in tadpoles results in a T cell differentiation and selection pathway distinct from that of post-metamorphic animals and relies more heavily on non-polymorphic nonclassical MHC class Ib (class Ib) molecules. Indeed, in the absence of optimal class Ia-mediated T cell selection, larval CD8 T cells may possess a more restricted antigen-binding repertoire, possibly reflected in the relative susceptibility of tadpoles to ranaviruses. However, as discussed later in this section, there are distinct class Ib-mediated T cell selection mechanisms and T cell subsets that may complement conventional class Ia-restricted CD8 T cells in tadpoles.
In contrast to tadpoles, adult X. laevis display conventional class Ia-restricted CD8 cytotoxic T cell populations. Despite the absence of available antibodies, CD4 T helper cells are also likely present owing to the presence of all genes involved in differentiation and function of CD4 T cells, the expression of the CD4 gene in CD8−/CD5+ cells, and the MHC class II-dependent proliferation response obtained by mixed lymphocyte reaction (Du Pasquier et al. 1989). The requirement of T cells for FV3 clearance in X. laevis adults has been demonstrated by using sub-lethal γ-irradiation, which depletes thymus-derived T cells. Irradiated T cell-depleted adult frogs do not control FV3 and succumb to infection (Robert et al. 2005). Furthermore, depletion of X. laevis CD8 T cells by administration of anti-X. laevis CD8 mAb also substantially increases adult frog susceptibility to FV3 infection (Robert et al. 2005). These CD8 T cell-depleted, FV3-infected animals experienced severe edema and hemorrhaging, extensive elevation of viral loads and succumbed to infections, whereas control cohorts effectively cleared the virus (Robert et al. 2005). Thus, cytotoxic CD8 T cell responses are critical for effective FV3 clearance. Intriguingly, administration of the anti-CD8 Ab to tadpoles did not result in either CD8 T cell depletion or in increased susceptibility to FV3 (Robert et al. 2005), again emphasizing the unconventional nature of the tadpole T cell populations.
Frogs re-infected with FV3 exhibit expedited viral clearance concomitant with earlier proliferation of CD5+CD8+ splenocytes and faster infiltration (3 vs. 6 dpi) of the kidney, the central site of X. laevis-FV3 replication (Morales and Robert 2007). This not only underlines the importance of CD8 T cells in ranaviral clearance, but also indicates the presence of a T cell memory responses to ranavirus re-infections in adult X. laevis. Interestingly, while kidney infiltration was substantially accelerated upon secondary FV3 challenge compared to primary infections, the numbers of recruited CD8+ cells were substantially lower during this second immune event (Morales and Robert 2007). This could be attributed to a higher frequency of T cell precursor infiltration upon the primary response and/or the generation of lowers number of more effective CD8+ memory T cells upon re-infection. It cannot be excluded that this modest secondary response is an inherent property of the evolutionarily primordial amphibian adaptive immune system, considering the relatively meager degree of T cell expansion seen following immunological challenges, the absence of draining lymph nodes and the lack of white pulp-red pulp splenic organization (Du Pasquier et al. 1989). Alternatively, this modest secondary CD8 response could be accounted for by the recruitment and immune involvement of additional effector populations during subsequent anti-ranavirus responses. In support of this notion, during the secondary anti-FV3 response, there is the rapid and robust recruitment, and kidney infiltration of, CD8− MHCII+ immune populations, which may be B cells, CD4 T cells, or CD8− nonclassical MHC (class Ib)-restricted invariant T cell populations.
5.3 Roles of Nonclassical MHC-Restricted Cells in Ranavirus Immunity
Nonclassical MHC class Ib (Ib) molecules exhibit structural similarities to class Ia molecules, but typically possess limited tissue distribution and substantially fewer polymorphisms (Flajnik and Kasahara 2001). In mammals, some of these surface glycoproteins are involved in the differentiation and functional regulation of distinct subsets of invariant T (iT) cells, including CD1d-restricted iNKT cells and MR1 restricted mucosal associated iT (MAIT) cells (Bendelac et al. 1995, 1996, 1997; Matsuda and Gapin 2005). Both of these lymphocyte populations undergo unconventional differentiation pathways, exhibit unique semi-invariant T cell receptor rearrangements and are believed to participate in antimicrobial and antiviral immune responses (Behar and Porcelli 2007; Choi et al. 2008; Cohen et al. 2009; Le Bourhis et al. 2010).
It is intriguing that, as described above, while Xenopus larvae are naturally MHC class Ia deficient (Du Pasquier et al. 1989), they express a number of nonclassical class Ib genes (XNCs), with some of these, such as XNC10 displaying preferential thymic expression (Goyos et al. 2009, 2011). Notably, we have recently identified a prominent X. laevis iT immune cell subset, which requires XNC10 for both its development and function (Edholm et al. 2013). Using an XNC10 tetramer as well as reverse genetics combining transgenesis and RNA interference, we determined that this iT cell population is CD8−/CD4−, expresses a semi-invariant T cell receptor consisting of an invariant TCRα (iVα6-Jα1.43) combined with a limited TCRβ repertoire, and fails to develop in the absence of, or with diminished, XNC10 expression (Edholm et al. 2013). Notably, transgenic animals with effectively RNAi-silenced thymic and splenic XNC10 expression failed to develop this iT cell subset. Moreover, they were also significantly more susceptible to, and more readily succumbed to, FV3 infections (Edholm et al. 2013) suggesting that these cells are important in anti-ranaviral defenses. It is noteworthy that deep sequencing analysis of tadpole TCRα revealed that Xenopus larvae possess several additional predominant iT cell populations (Edholm et al. 2013), which are presumably XNC-restricted and most likely participate in immune responses such as those against ranaviruses. Indeed, we have also identified the XNC10-dependent iT cell population in adult frogs; thus it stands to reason that during the primary and secondary anti-FV3 responses, these lymphocyte subsets may be amongst the CD8− kidney infiltrating immune populations (Morales and Robert 2007) discussed above. Table 1 provides a comprehensive summary of host immune strategies.
6 Ranaviral Strategies for Evading Host Antiviral Immunity
As seen with poxviruses and other large DNA viruses, ranaviruses likely encode multiple proteins that function to impede the host antiviral response (Finlay and McFadden 2006; Johnston and McFadden 2003; Seet et al. 2003). However, with the exception of the ranavirus homolog of the largest subunit of eukaryotic initiation factor 2 (vIF-2α), the functions of these gene products have not been determined. A description of the role of vIF-2α follows, along with a brief description of other potential immune evasion proteins.
6.1 vIF-2α Blocks Phosphorylation of eIF-2α
Protein kinase R (PKR, EIF2αK2) is a protein kinase that regulates cellular protein synthesis via phosphorylation and inactivation of the alpha subunit of eukaryotic translation initiation factor 2 (eIF2α) in response to a variety of environmental stressors including viral infection (Proud 1995; Toth et al. 2006). PKR is present at low levels within uninfected cells and is induced following cell stimulation by antiviral IFNs. PKR is an inactive monomer in uninfected cells. However, during viral infection, low concentrations of viral dsRNA bind PKR leading to its dimerization and activation via autophosphorylation (Zhang et al. 2001). Activated PKR subsequently phosphorylates the alpha subunit of eIF-2, an event that results in a global arrest in protein synthesis (Panniers et al. 1988; Rowlands et al. 1988). In addition to the effect of PKR-mediated eIF-2α inactivation on protein synthesis, activated PKR may play additional roles. Activated PKR phosphorylates an inhibitor, I-κB, bound to NF-κB leading to the latter’s release and the subsequent activation of pro-inflammatory and interferon genes (Proud 1995). In addition, activated PKR appears to be one of the danger signals that trigger apoptosis in virus infected cells. Because of the adverse effects that translational inhibition, NF-κB activation, and apoptosis have on virus replication, viruses have evolved numerous approaches for circumventing PKR-mediated antiviral functions (Diener et al. 1993; Katze 1992).
To block phosphorylation of eIF-2α, ranaviruses encode a pseudosubstrate of eIF-2α designated vIF-2α. In most ranaviruses, vIF-2α is present as protein, of about 250 amino acid residues in length, that contains a sequence motif (V[L/I]RVDxxKGY[V/I]D) common to multiple ranaviruses, host cell eIF-2α, and the K3L protein of vaccinia virus (Majji et al. 2006). K3L has been shown to block the phosphorylation and subsequent inactivation of eIF-2α by acting as a pseudosubstrate for PKR (Beattie et al. 1995), and vIF-2α has been shown to function in similar fashion (see below). By using an ATV KO mutant lacking vIF-2α, Jancovich and Jacobs (Jancovich and Jacobs 2011) showed that this mutant virus was more sensitive to IFN-mediated inhibition and failed to block the phosphorylation of eIF-2α. Moreover, whereas wt ATV degraded fish PKZ, an IFN-inducible kinase similar to PKR, the KO mutant did not. Lastly, the KO mutant displayed reduced virulence in vivo suggesting that vIF-2α is a virulence gene. A similar attenuation of virulence was observed following infection of Xenopus laevis with a FV3 KO mutant lacking a truncated version of vIF-2α (Chen et al. 2011). Truncated versions of vIF-2α are found in FV3, soft-shell turtle iridovirus, and Rana grylio virus and are missing the N-terminal half of the native molecule. Since this region contains the VxRVDxxKGYxD motif described above, the reduction in virulence cannot be due to an effect of vIF-2α on PKR, but to some element present within the C-terminal half of the protein.
Rothenburg et al. (2011), using a yeast model, demonstrated that transfection of a vector expressing either human or zebrafish PKR into yeast cells resulted in marked cell death. Furthermore, in confirmation of the role of vIF-2α as an antagonist of PKR, co-transfection of a vector expressing the full-length vIF-2α gene from Rana catesbeiana virus (RCV) along with PKR blocked the toxic effects of both human and zebrafish PKR. Indicative of species specificity, vaccinia virus K3L was only able to block the activity of human PKR. Although the above study indicates a role for vIF-2α in maintaining protein synthesis in virus-infected cells, the observation that FV3 and other closely related ranaviruses contain truncated vIF-2α genes lacking critical N-terminal motifs indicates that vIF-2α may not be the only ranavirus protein that plays a role in maintaining protein synthesis in virus infected cells.
6.2 RNAse III-Like Proteins
Similar to poxviruses, ranaviruses may also contain at least two genes whose function is to prevent the activation of PKR. Vaccinia virus encodes both the aforementioned K3L gene and a second gene, E3L, which binds dsRNA and prevents the dimerization and activation of PKR (Langland and Jacobs 2002; Langland et al. 2006). Although no ranavirus protein with homology to E3L has been detected, an RNAse III-like protein has been identified. RNAse-III targets dsRNA and it is possible that ranavirus homologs bind virus-induced dsRNA and degrade it, or block its ability to interact with and activate PKR. Experiments to directly test this hypothesis have not been reported. Moreover, knock down of RNAse III-like protein expression using antisense morpholino oligonucleotides resulted in a 40% reduction in virus yield (K. Cheng and V.G. Chinchar, University of Mississippi, unpublished data) suggesting that the RNaseIII-like protein plays a role in virus replication.
6.3 β-Hydroxysteroid Dehydrogenase and vCARD
Ranaviruses, like poxviruses, contain proteins with homology to β-hydroxysteroid dehydrogenase (βHSD). βHSD plays a role in steroid synthesis and expression of a βHSD homolog by vaccinia virus results in suppression of immunity and an increase in viral replication (Sroller et al. 1998). vCARD is a 10 kDa, virus-encoded protein that contains a Caspase Activation and Recruitment Domain (CARD) motif that modulates interaction between proteins bearing similar domains (Kawai and Akira 2009, 2010). Because proteins involved in apoptosis or in the induction of IFN and pro-inflammatory molecules such as RIG-I, MDA5, and MAVS contain CARD motifs, it is postulated that vCARD interacts with one or more of these signaling molecules and short-circuits cellular antiviral immunity (Besch et al. 2009; Meylan et al. 2005).
6.4 vTNFR, dUTPase, DMTase
In addition to the four viral gene products mentioned above, ranaviruses also contain homologs of Tumor Necrosis Factor (TNF) receptor (vTNFR) and dUTPase (Chinchar et al. 2009; Eaton et al. 2007) and a unique virus-encoded DNA cytosine methyltransferase (DMTase). Similar to their poxvirus counterparts, ranavirus vTNFR could function as a decoy molecule and block protection mediated by TNFα. Although dUTPase is generally considered to be a protein that plays a role in viral DNA synthesis (e.g., by increasing dTTP pools or blocking the incorporation of dUTP into DNA), a herpesvirus dUTPase was shown to also block antiviral immunity (Glaser et al. 2006; Oliveros et al. 1999). Lastly, the ranavirus DMTase may play a role in immune evasion by methylating cytosine residues within CpG motifs and blocking recognition by TLR-9 or cytoplasmic DNA sensors and preventing the subsequent induction of IFN and pro-inflammatory cytokines (Krieg 2002; Krug et al. 2004).
The above list of putative immune evasion proteins is based on ranavirus proteins with detectable homology to proteins in other systems with known antiviral effects. While this serves as a useful starting point in the identification and characterization of ranaviral immune evasion genes, there are approximately a dozen additional ORFs of unknown function, which are unique to ranaviruses. Whether these ORFs encode proteins that control virus replication in specific hosts or whether they encode proteins that modulate host-specific immune responses remains to be determined. Knock-down experiments using antisense morpholino oligonucleotides or siRNA and infections using knock-out mutants will be needed to resolve the function of these unique ranavirus-specific proteins.
7 Concluding Remarks and Future Directions
It is evident from the studies described here that anti-ranaviral immunity is multifaceted, complex, and likely species- and developmental stage-specific. Also evident are the many gaps in our understanding of the immune response to these pathogens as well as possible defects in the host’s ability to mount effective responses that contain and eliminate these infections. It is particularly worrisome that Ranavirus and other genera within the family Iridoviridae have devised numerous, highly efficient strategies for evading, and even utilizing host immune components, to achieve persistence, facilitate dissemination and expand host range. Clearly, ranaviruses encode a large number of putative gene products, which represent both potential virulence factors as well as promising targets for future therapeutic interventions.
While it is easy to dismiss lower vertebrate immune systems as functionally analogous to those of mammals, there is a growing literature suggesting otherwise. It is through the fundamental understanding of the physiological and ecological pressures governing these unique immune systems that we may begin to comprehend ranavirus infection strategies and the immune systems that may, or may not have adequately co-evolved to stop them.
The investigation of ranavirus infection and immune subversion strategies should be approached not only by taking into account well-defined mammalian pathogens, but also by considering the possibility that ranaviruses may represent unique viral agents. In contrast to the majority of homeothermic vertebrate pathogens, ranaviruses are extraordinary in their ability to overcome cell and host tropism barriers, while their mechanisms of pathogenicity appear to be (at least partially) much less dependent on viral loads. Indeed, the immune systems of ectothermic hosts have evolved as the result of, and are subject to, different physiological and pathogenic pressures than those that have shaped the mammalian immune system. It stands to reason that ranavirus pathogens have co-evolved with these unique immune systems, thus we must garner greater insights into both to fully understand either.
References
Aggad D, Mazel M, Boudinot P et al (2009) The two groups of zebrafish virus-induced interferons signal via distinct receptors with specific and shared chains. J Immunol 183:3924–3931
Altmann SM, Mellon MT, Distel DL et al (2003) Molecular and functional analysis of an interferon gene from the zebrafish, Danio rerio. J Virol 77:1992–2002
Alvarez-Torres D, Garcia-Rosado E, Fernandez-Trujillo MA et al (2013) Antiviral specificity of the Solea senegalensis Mx protein constitutively expressed in CHSE-214 cells. Mar Biotechnol (NY) 15:125–132
Aubertin AM, Hirth C, Travo C et al (1973) Preparation and properties of an inhibitory extract from frog virus 3 particles. J Virol 11:694–701
Auffray C, Fogg D, Garfa M et al (2007) Monitoring of blood vessels and tissues by a population of monocytes with patrolling behavior. Science 317:666–670
Baum A, Garcia-Sastre A (2010) Induction of type I interferon by RNA viruses: cellular receptors and their substrates. Amino Acids 38:1283–1299
Bayley AE, Hill BJ, Feist SW (2013) Susceptibility of the European common frog Rana temporaria to a panel of ranavirus isolates from fish and amphibian hosts. Dis Aquat Organ 103:171–183
Beattie E, Paoletti E, Tartaglia J (1995) Distinct patterns of IFN sensitivity observed in cells infected with vaccinia K3L- and E3L-mutant viruses. Virology 210:254–263
Behar SM, Porcelli SA (2007) CD1-restricted T cells in host defense to infectious diseases. Curr Top Microbiol Immunol 314:215–250
Behncke H, Stohr AC, Heckers KO et al (2013) Mass-mortality in green striped tree dragons (Japalura splendida) associated with multiple viral infections. Vet Rec 173:248
Bendelac A, Lantz O, Quimby ME et al (1995) CD1 recognition by mouse NK1+ T lymphocytes. Science 268:863–865
Bendelac A, Hunziker RD, Lantz O (1996) Increased interleukin 4 and immunoglobulin E production in transgenic mice overexpressing NK1 T cells. J Exp Med 184:1285–1293
Bendelac A, Rivera MN, Park SH et al (1997) Mouse CD1-specific NK1 T cells: development, specificity, and function. Annu Rev Immunol 15:535–562
Besch R, Poeck H, Hohenauer T et al (2009) Proapoptotic signaling induced by RIG-I and MDA-5 results in type I interferon-independent apoptosis in human melanoma cells. J Clin Invest 119:2399–2411
Blomberg B, Bernard CC, Du Pasquier L (1980) In vitro evidence for T-B lymphocyte collaboration in the clawed toad, Xenopus. Eur J Immunol 10:869–876
Bollinger TK, Mao J, Schock D et al (1999) Pathology, isolation, and preliminary molecular characterization of a novel iridovirus from tiger salamanders in Saskatchewan. J Wildl Dis 35:413–429
Brunner JL, Schock DM, Davidson EW et al (2004) Intraspective reservoires: complex life hoistory and the persistance of a lethal ranavirus. Ecology 85:560–566
Caipang CM, Hirono I, Aoki T (2003) In vitro inhibition of fish rhabdoviruses by Japanese flounder, Paralichthys olivaceus Mx. Virology 317:373–382
Caipang CM, Takano T, Hirono I et al (2006) Genetic vaccines protect red seabream, Pagrus major, upon challenge with red seabream iridovirus (RSIV). Fish Shellfish Immunol 21:130–138
Cano I, Alonso MC, Garcia-Rosado E et al (2006) Detection of lymphocystis disease virus (LCDV) in asymptomatic cultured gilt-head seabream (Sparus aurata, L.) using an immunoblot technique. Vet Microbiol 113:137–141
Cano I, Ferro P, Alonso MC et al (2009) Application of in situ detection techniques to determine the systemic condition of lymphocystis disease virus infection in cultured gilt-head seabream, Sparus aurata L. J Fish Dis 32:143–150
Carey C, Cohen N, Rollins-Smith L (1999) Amphibian declines: an immunological perspective. Dev Comp Immunol 23:459–472
Chang M, Nie P, Collet B et al (2009) Identification of an additional two-cysteine containing type I interferon in rainbow trout Oncorhynchus mykiss provides evidence of a major gene duplication event within this gene family in teleosts. Immunogenetics 61:315–325
Chao CB, Chen CY, Lai YY et al (2004) Histological, ultrastructural, and in situ hybridization study on enlarged cells in grouper Epinephelus hybrids infected by grouper iridovirus in Taiwan (TGIV). Dis Aquat Organ 58:127–142
Chen G, Robert J (2011) Antiviral immunity in amphibians. Viruses 3:2065–2086
Chen G, Ward BM, Yu KH et al (2011) Improved knockout methodology reveals that frog virus 3 mutants lacking either the 18K immediate-early gene or the truncated vIF-2alpha gene are defective for replication and growth in vivo. J Virol 85:11131–11138
Cheng K, Escalon BL, Robert J et al (2014) Differential transcription of fathead minnow immune-related genes following infection with frog virus 3, an emerging pathogen of ectothermic vertebrates. Virology 456–457:77–86
Chinchar VG, Wang J, Murti G et al (2001) Inactivation of frog virus 3 and channel catfish virus by esculentin-2P and ranatuerin-2P, two antimicrobial peptides isolated from frog skin. Virology 288:351–357
Chinchar VG, Bryan L, Wang J et al (2003) Induction of apoptosis in frog virus 3-infected cells. Virology 306:303–312
Chinchar VG, Hyatt A, Miyazaki T et al (2009) Family Iridoviridae: poor viral relations no longer. Curr Top Microbiol Immunol 328:123–170
Choi HJ, Xu H, Geng Y et al (2008) Bacterial infection alters the kinetics and function of iNKT cell responses. J Leukoc Biol 84:1462–1471
Chung SW, Arnott JA, Yang Y et al (2003) Presence of prepackaged mRNA in virions of DNA adenovirus. J Biol Chem 278:50635–50640
Cohen NR, Garg S, Brenner MB (2009) Antigen presentation by CD1 lipids, T cells, and NKT cells in microbial immunity. Adv Immunol 102:1–94
Coiras M, Lopez-Huertas MR, Perez-Olmeda M et al (2009) Understanding HIV-1 latency provides clues for the eradication of long-term reservoirs. Nat Rev Microbiol 7:798–812
Cotter JD, Storfer A, Page RB et al (2008) Transcriptional response of Mexican axolotls to Ambystoma tigrinum virus (ATV) infection. BMC Genomics 9:493
De Jesús Andino F, Chen G, Li Z et al (2012) Susceptibility of Xenopus laevis tadpoles to infection by the ranavirus Frog-Virus 3 correlates with a reduced and delayed innate immune response in comparison with adult frogs. Virology 432:435–443
Diener TO, Hammond RW, Black T et al (1993) Mechanism of viroid pathogenesis: differential activation of the interferon-induced, double-stranded RNA-activated, M(r) 68,000 protein kinase by viroid strains of varying pathogenicity. Biochimie 75:533–538
Du Pasquier L, Schwager J, Flajnik MF (1989) The immune system of Xenopus. Annu Rev Immunol 7:251–275
Du Pasquier L, Robert J, Courtet M et al (2000) B-cell development in the amphibian Xenopus. Immunol Rev 175:201–213
Duffus ALJ, Nichols RA, Garner TWJ (2013) Investigations into the life history stages of the common frog (Rana temporaria) affected by an amphibian ranavirus in the United Kingdom. Herpetol Rev 44:260–263
Eaton HE, Metcalf J, Penny E et al (2007) Comparative genomic analysis of the family Iridoviridae: re-annotating and defining the core set of iridovirus genes. Virol J 4:11
Edholm ES, Albertorio Saez LM, Gill AL et al (2013) Nonclassical MHC class I-dependent invariant T cells are evolutionarily conserved and prominent from early development in amphibians. Proc Natl Acad Sci U S A 110:14342–14347
Elharrar M, Hirth C, Blanc J et al (1973) Pathogenesis of the toxic hepatitis of mice provoked by FV 3 (frog virus 3): inhibition of liver macromolecular synthesis (author’s transl). Biochim Biophys Acta 319:91–102
Fernandez-Trujillo MA, Garcia-Rosado E, Alonso MC et al (2013) Mx1, Mx2 and Mx3 proteins from the gilthead seabream (Sparus aurata) show in vitro antiviral activity against RNA and DNA viruses. Mol Immunol 56:630–636
Finlay BB, McFadden G (2006) Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell 124:767–782
Flajnik MF, Kasahara M (2001) Comparative genomics of the MHC: glimpses into the evolution of the adaptive immune system. Immunity 15:351–362
Fullerton JN, O’Brien AJ, Gilroy DW (2013) Pathways mediating resolution of inflammation: when enough is too much. J Pathol 231:8–20
Gantress J, Maniero GD, Cohen N et al (2003) Development and characterization of a model system to study amphibian immune responses to iridoviruses. Virology 311:254–262
Gendrault JL, Steffan AM, Bingen A et al (1981) Penetration and uncoating of frog virus 3 (FV3) in cultured rat Kupffer cells. Virology 112:375–384
Glaser R, Litsky ML, Padgett DA et al (2006) EBV-encoded dUTPase induces immune dysregulation: implications for the pathophysiology of EBV-associated disease. Virology 346:205–218
Goodenow MM, Rose SL, Tuttle DL et al (2003) HIV-1 fitness and macrophages. J Leukoc Biol 74:657–666
Gousset K, Ablan SD, Coren LV et al (2008) Real-time visualization of HIV-1 GAG trafficking in infected macrophages. PLoS Pathog 4:e1000015
Goyos A, Ohta Y, Guselnikov S et al (2009) Novel nonclassical MHC class Ib genes associated with CD8 T cell development and thymic tumors. Mol Immunol 46:1775–1786
Goyos A, Sowa J, Ohta Y et al (2011) Remarkable conservation of distinct nonclassical MHC class I lineages in divergent amphibian species. J Immunol 186:372–381
Gray MJ, Miller DL (2013) Rise of ranavirus: an emerging pathogen threatens ectothermic vertebrates. Wildl Professional 7:51–55
Grayfer L, Garcia EG, Belosevic M (2010) Comparison of macrophage antimicrobial responses induced by type II interferons of the goldfish (Carassius auratus L.). J Biol Chem 285:23537–23547
Grayfer L, De Jesús Andino F, Robert J (2014) The amphibian (Xenopus laevis) type I interferon response to Frog Virus 3: new insight into ranavirus pathogenicity. J Virol 88(10):5766–5777
Groot F, Welsch S, Sattentau QJ (2008) Efficient HIV-1 transmission from macrophages to T cells across transient virological synapses. Blood 111:4660–4663
Gut JP, Anton M, Bingen A et al (1981) Frog virus 3 induces a fatal hepatitis in rats. Lab Invest 45:218–228
Hagmann W, Steffan AM, Kirn A et al (1987) Leukotrienes as mediators in frog virus 3-induced hepatitis in rats. Hepatology 7:732–736
Haislip NA, Gray MJ, Hoverman JT et al (2011) Development and disease: how susceptibility to an emerging pathogen changes through anuran development. PLoS One 6:e22307
Hervas-Stubbs S, Perez-Gracia JL, Rouzaut A et al (2011) Direct effects of type I interferons on cells of the immune system. Clin Cancer Res 17:2619–2627
Holopainen R, Tapiovaara H, Honkanen J (2012) Expression analysis of immune response genes in fish epithelial cells following ranavirus infection. Fish Shellfish Immunol 32:1095–1105
Horton JD, Horton TL, Dzialo R et al (1998) T-cell and natural killer cell development in thymectomized Xenopus. Immunol Rev 166:245–258
Hoverman JT, Gray MJ, Miller DL (2010) Anuran susceptibilities to ranaviruses: role of species identity, exposure route, and a novel virus isolate. Dis Aquat Organ 89:97–107
Hsu E (1998) Mutation, selection, and memory in B lymphocytes of exothermic vertebrates. Immunol Rev 162:25–36
Hyatt AD, Williamson M, Coupar BE et al (2002) First identification of a ranavirus from green pythons (Chondropython viridis). J Wildl Dis 38:239–252
Jancovich JK, Jacobs BL (2011) Innate immune evasion mediated by the Ambystoma tigrinum virus eukaryotic translation initiation factor 2alpha homologue. J Virol 85:5061–5069
Joerink M, Forlenza M, Ribeiro CM et al (2006a) Differential macrophage polarisation during parasitic infections in common carp (Cyprinus carpio L.). Fish Shellfish Immunol 21:561–571
Joerink M, Ribeiro CM, Stet RJ et al (2006b) Head kidney-derived macrophages of common carp (Cyprinus carpio L.) show plasticity and functional polarization upon differential stimulation. J Immunol 177:61–69
Joerink M, Savelkoul HF, Wiegertjes GF (2006c) Evolutionary conservation of alternative activation of macrophages: structural and functional characterization of arginase 1 and 2 in carp (Cyprinus carpio L.). Mol Immunol 43:1116–1128
Johnston JB, McFadden G (2003) Poxvirus immunomodulatory strategies: current perspectives. J Virol 77:6093–6100
Jurgens JB, Gartland LA, Du Pasquier L et al (1995) Identification of a candidate CD5 homologue in the amphibian Xenopus laevis. J Immunol 155:4218–4223
Katze MG (1992) The war against the interferon-induced dsRNA-activated protein kinase: can viruses win? J Interferon Res 12:241–248
Kawai T, Akira S (2009) The roles of TLRs, RLRs and NLRs in pathogen recognition. Int Immunol 21:317–337
Kawai T, Akira S (2010) The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol 11:373–384
Kirn A, Gut JP, Elharrar M (1972) [FV3 (Frog Virus 3) toxicity for the mouse]. Nouv Presse Med 1:1943
Kirn A, Steffan AM, Bingen A (1980) Inhibition of erythrophagocytosis by cultured rat Kupffer cells infected with frog virus 3. J Reticuloendothel Soc 28:381–388
Kirn A, Bingen A, Steffan AM et al (1982) Endocytic capacities of Kupffer cells isolated from the human adult liver. Hepatology 2:216–222
Kochs G, Reichelt M, Danino D et al (2005) Assay and functional analysis of dynamin-like Mx proteins. Methods Enzymol 404:632–643
Kotenko SV (2011) IFN-lambdas. Curr Opin Immunol 23:583–590
Krieg AM (2002) CpG motifs in bacterial DNA and their immune effects. Annu Rev Immunol 20:709–760
Krug LT, Pozharskaya VP, Yu Y et al (2004) Inhibition of infection and replication of human herpesvirus 8 in microvascular endothelial cells by alpha interferon and phosphonoformic acid. J Virol 78:8359–8371
Landsberg JH, Kiryu Y, Tabuchi M et al (2013) Co-infection by alveolate parasites and frog virus 3-like ranavirus during an amphibian larval mortality event in Florida, USA. Dis Aquat Organ 105:89–99
Langland JO, Jacobs BL (2002) The role of the PKR-inhibitory genes, E3L and K3L, in determining vaccinia virus host range. Virology 299:133–141
Langland JO, Kash JC, Carter V et al (2006) Suppression of proinflammatory signal transduction and gene expression by the dual nucleic acid binding domains of the vaccinia virus E3L proteins. J Virol 80:10083–10095
Le Bourhis L, Martin E, Peguillet I et al (2010) Antimicrobial activity of mucosal-associated invariant T cells. Nat Immunol 11:701–708
Leibovitz L (1980) Lymphocystis disease. J Am Vet Med Assoc 176:202
Lester K, Hall M, Urquhart K et al (2012) Development of an in vitro system to measure the sensitivity to the antiviral Mx protein of fish viruses. J Virol Methods 182:1–8
Levraud JP, Boudinot P, Colin I et al (2007) Identification of the zebrafish IFN receptor: implications for the origin of the vertebrate IFN system. J Immunol 178:4385–4394
Li Z, Strunk JJ, Lamken P et al (2008) The EM structure of a type I interferon-receptor complex reveals a novel mechanism for cytokine signaling. J Mol Biol 377:715–724
Li Z, Xu X, Huang L et al (2010) Administration of recombinant IFN1 protects zebrafish (Danio rerio) from ISKNV infection. Fish Shellfish Immunol 29:399–406
Long S, Wilson M, Bengten E et al (2004) Identification of a cDNA encoding channel catfish interferon. Dev Comp Immunol 28:97–111
Lopez C, Aubertin AM, Tondre L et al (1986) Thermosensitivity of frog virus 3 genome expression: defect in early transcripti. Virology 152:365–374
Lopez-Munoz A, Roca FJ, Meseguer J et al (2009) New insights into the evolution of IFNs: zebrafish group II IFNs induce a rapid and transient expression of IFN-dependent genes and display powerful antiviral activities. J Immunol 182:3440–3449
Lutfalla G, Roest Crollius H, Stange-Thomann N et al (2003) Comparative genomic analysis reveals independent expansion of a lineage-specific gene family in vertebrates: the class II cytokine receptors and their ligands in mammals and fish. BMC Genomics 4:29
Majji S, LaPatra S, Long SM et al (2006) Rana catesbeiana virus Z (RCV-Z): a novel pathogenic ranavirus. Dis Aquat Organ 73:1–11
Maniero GD, Morales H, Gantress J et al (2006) Generation of a long-lasting, protective, and neutralizing antibody response to the ranavirus FV3 by the frog Xenopus. Dev Comp Immunol 30:649–657
Matsuda JL, Gapin L (2005) Developmental program of mouse Valpha14i NKT cells. Curr Opin Immunol 17:122–130
Meylan E, Curran J, Hofmann K et al (2005) Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167–1172
Miller D, Gray M, Storfer A (2011) Ecopathology of ranaviruses infecting amphibians. Viruses 3:2351–2373
Morales HD, Robert J (2007) Characterization of primary and memory CD8 T-cell responses against ranavirus (FV3) in Xenopus laevis. J Virol 81:2240–2248
Morales HD, Abramowitz L, Gertz J et al (2010) Innate immune responses and permissiveness to ranavirus infection of peritoneal leukocytes in the frog Xenopus laevis. J Virol 84:4912–4922
Nahrendorf M, Swirski FK, Aikawa E et al (2007) The healing myocardium sequentially mobilizes two monocyte subsets with divergent and complementary functions. J Exp Med 204:3037–3047
Nakajima K, Kunita J (2005) [Red sea bream iridoviral disease]. Uirusu 55:115–125
Nakajima K, Maeno Y, Honda A et al (1999) Effectiveness of a vaccine against red sea bream iridoviral disease in a field trial test. Dis Aquat Organ 36:73–75
Nishikawa A, Murata E, Akita M et al (1998) Roles of macrophages in programmed cell death and remodeling of tail and body muscle of Xenopus laevis during metamorphosis. Histochem Cell Biol 109:11–17
Oliveros M, Garcia-Escudero R, Alejo A et al (1999) African swine fever virus dUTPase is a highly specific enzyme required for efficient replication in swine macrophages. J Virol 73:8934–8943
Panniers R, Rowlands AG, Henshaw EC (1988) The effect of Mg2+ and guanine nucleotide exchange factor on the binding of guanine nucleotides to eukaryotic initiation factor 2. J Biol Chem 263:5519–5525
Pham PH, Lai YS, Lee FF et al (2012) Differential viral propagation and induction of apoptosis by grouper iridovirus (GIV) in cell lines from three non-host species. Virus Res 167:16–25
Proud CG (1995) PKR: a new name and new roles. Trends Biochem Sci 20:241–246
Qi Z, Nie P, Secombes CJ et al (2010) Intron-containing type I and type III IFN coexist in amphibians: refuting the concept that a retroposition event gave rise to type I IFNs. J Immunol 184:5038–5046
Raghow R, Granoff A (1979) Macromolecular synthesis in cells infected by frog virus 3. X. Inhibition of cellular protein synthesis by heat-inactivated virus. Virology 98:319–327
Reeve BC, Crespi EJ, Whipps CM et al (2013) Natural stressors and ranavirus susceptibility in larval wood frogs (Rana sylvatica). Ecohealth 10:190–200
Robert J, Ohta Y (2009) Comparative and developmental study of the immune system in Xenopus. Dev Dyn 238:1249–1270
Robert J, Morales H, Buck W et al (2005) Adaptive immunity and histopathology in frog virus 3-infected Xenopus. Virology 332:667–675
Robert J, Abramowitz L, Gantress J et al (2007) Xenopus laevis: a possible vector of Ranavirus infection? J Wildl Dis 43:645–652
Robert J, George E, De Jesús Andino F et al (2011) Waterborne infectivity of the Ranavirus frog virus 3 in Xenopus laevis. Virology 417:410–417
Robertsen B (2006) The interferon system of teleost fish. Fish Shellfish Immunol 20:172–191
Robertsen B, Bergan V, Rokenes T et al (2003) Atlantic salmon interferon genes: cloning, sequence analysis, expression, and biological activity. J Interferon Cytokine Res 23:601–612
Rollins-Smith LA (2009) The role of amphibian antimicrobial peptides in protection of amphibians from pathogens linked to global amphibian declines. Biochim Biophys Acta 1788:1593–1599
Rollins-Smith LA, Reinert LK, O’Leary CJ et al (2005) Antimicrobial peptide defenses in amphibian skin. Integr Comp Biol 45:137–142
Rothenburg S, Chinchar VG, Dever TE (2011) Characterization of a ranavirus inhibitor of the antiviral protein kinase PKR. BMC Microbiol 11:56
Rowlands AG, Panniers R, Henshaw EC (1988) The catalytic mechanism of guanine nucleotide exchange factor action and competitive inhibition by phosphorylated eukaryotic initiation factor 2. J Biol Chem 263:5526–5533
Sadler AJ, Williams BR (2008) Interferon-inducible antiviral effectors. Nat Rev Immunol 8:559–568
Samuel CE (2001) Antiviral actions of interferons. Clin Microbiol Rev 14:778–809, Table of contents
Seet BT, Johnston JB, Brunetti CR et al (2003) Poxviruses and immune evasion. Annu Rev Immunol 21:377–423
Siwicki AK, Pozet F, Morand M et al (1999) Effects of iridovirus-like agent on the cell-mediated immunity in sheatfish (Silurus glanis)—an in vitro study. Virus Res 63:115–119
Sroller V, Kutinova L, Nemeckova S et al (1998) Effect of 3-beta-hydroxysteroid dehydrogenase gene deletion on virulence and immunogenicity of different vaccinia viruses and their recombinants. Arch Virol 143:1311–1320
Sun B, Robertsen B, Wang Z et al (2009) Identification of an Atlantic salmon IFN multigene cluster encoding three IFN subtypes with very different expression properties. Dev Comp Immunol 33:547–558
Sutton WB, Gray MJ, Hardman RH et al (2014) High susceptibility of the endangered dusky gopher frog to ranavirus. Dis Aquat Organ 112(1):9–16
Svingerud T, Solstad T, Sun B et al (2012) Atlantic salmon type I IFN subtypes show differences in antiviral activity and cell-dependent expression: evidence for high IFNb/IFNc-producing cells in fish lymphoid tissues. J Immunol 189:5912–5923
Toth AM, Zhang P, Das S et al (2006) Interferon action and the double-stranded RNA-dependent enzymes ADAR1 adenosine deaminase and PKR protein kinase. Prog Nucleic Acid Res Mol Biol 81:369–434
Trobridge GD, Chiou PP, Leong JA (1997) Cloning of the rainbow trout (Oncorhynchus mykiss) Mx2 and Mx3 cDNAs and characterization of trout Mx protein expression in salmon cells. J Virol 71:5304–5311
Turner RJ, Manning MJ (1974) Thymic dependence of amphibian antibody responses. Eur J Immunol 4:343–346
Wiegertjes GF, Forlenza M (2010) Nitrosative stress during infection-induced inflammation in fish: lessons from a host-parasite infection model. Curr Pharm Des 16:4194–4202
Wu YC, Chi SC (2007) Cloning and analysis of antiviral activity of a barramundi (Lates calcarifer) Mx gene. Fish Shellfish Immunol 23:97–108
Wu MS, Chen CW, Liu YC et al (2012) Transcriptional analysis of orange-spotted grouper reacting to experimental grouper iridovirus infection. Dev Comp Immunol 37:233–242
Yang SJ, Xiao XH, Xu YG et al (2012) Induction of antimicrobial peptides from Rana dybowskii under Rana grylio virus stress, and bioactivity analysis. Can J Microbiol 58:848–855
Zhang F, Romano PR, Nagamura-Inoue T et al (2001) Binding of double-stranded RNA to protein kinase PKR is required for dimerization and promotes critical autophosphorylation events in the activation loop. J Biol Chem 276:24946–24958
Zhao C, Zhang H, Wong WC et al (2009) Identification of novel functional differences in monocyte subsets using proteomic and transcriptomic methods. J Proteome Res 8:4028–4038
Zhu R, Wang J, Lei XY et al (2013) Evidence for Paralichthys olivaceus IFITM1 antiviral effect by impeding viral entry into target cells. Fish Shellfish Immunol 35:918–926
Ziegler-Heitbrock L (2007) The CD14+ CD16+ blood monocytes: their role in infection and inflammation. J Leukoc Biol 81:584–592
Zilberg D, Grizzle JM, Plumb JA (2000) Preliminary description of lesions in juvenile largemouth bass injected with largemouth bass virus. Dis Aquat Organ 39:143–146
Zou J, Secombes CJ (2011) Teleost fish interferons and their role in immunity. Dev Comp Immunol 35:1376–1387
Zou J, Tafalla C, Truckle J et al (2007) Identification of a second group of type I IFNs in fish sheds light on IFN evolution in vertebrates. J Immunol 179:3859–3871
Acknowledgments
We thank Louise Rollins-Smith (Vanderbilt University) for reviewing an earlier draft of the manuscript. L. Grayfer was supported by postdoctoral fellowships from the Natural Sciences and Engineering Research Council of Canada and the Howard Hughes Medical Institute—Life Sciences Research Foundation. F. De Jesús Andino research support D14ZO-084 from the Morris Foundation. J. Robert and E.S. Edholm research support: 2 R24 Al 059830-10 from the NIH, and J. Robert and V.G. Chinchar IOS-0923772 and IOS-0742711 from the NSF.
Open Access publication was made possible through grants provided by the University of Tennessee (Institute of Agriculture, Office of Research and Engagement, and Department of Forestry, Wildlife and Fisheries), Washington State University Libraries, Gordon State College (Office of Academic Affairs), the Association of Reptilian and Amphibian Veterinarians, and the Amphibian and Reptile Conservancy.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is distributed under the terms of the Creative Commons Attribution Noncommercial License, which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Copyright information
© 2015 The Author(s)
About this chapter
Cite this chapter
Grayfer, L., Edholm, ES., De Jesús Andino, F., Chinchar, V.G., Robert, J. (2015). Ranavirus Host Immunity and Immune Evasion. In: Gray, M., Chinchar, V. (eds) Ranaviruses. Springer, Cham. https://doi.org/10.1007/978-3-319-13755-1_6
Download citation
DOI: https://doi.org/10.1007/978-3-319-13755-1_6
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-13754-4
Online ISBN: 978-3-319-13755-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)