Abstract
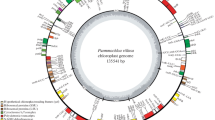
Orinus kokonoricus is an alpine perennial grass (Poaceae) endemic to the Qinghai-Tibet Plateau (QTP) in China, which has extremely important ecological and genetic values. To explore characterization and phylogenetic analysis of the complete chloroplast (cp) genome of O. kokonoricus, we first sequenced and assembled its cp genome with Illumina HiSeq4000 platform in the present study. The results showed that the complete cp genome of O. kokonoricus is 134,466 bp in length with a high AT content of 61.6% and displays a standard quadripartite structure including one large single copy region (LSC, 79,932 bp), one small single copy region (SSC, 12,490 bp) and two inverted repeat regions (IRA and IRB, 21,022 bp each). It totally encodes 137 genes containing 81 protein-coding genes, 45 tRNAs genes and eight rRNAs genes. Moreover, most of these genes occur in the single copy regions. Among all annotated genes of the cp genome of O. kokonoricus, none of them harbors introns. In addition, phylogenetic analysis based on 40 complete cp genome sequences of Poaceae revealed that O. kokonoricus is sister clade to the clade of Eragrostis species in Chloridoideae.


Similar content being viewed by others
REFERENCES
Gray, J.C., Genetic manipulation of the chloroplast genome, Biotechnology, 1989, vol. 12, no. 14, pp. 317–335.
Howe, C.J., Barbrook, A.C., Koumandou, V.L., Nisbet, R.E., and Symington, H.A., Evolution of the chloroplast genome, Philos. Trans. R. Soc. London, Ser. B: Biol. Sci., 2003, vol. 358, no. 1429, pp. 99–107.
Jansen, R.K., Cai, Z.Q., Raubeson, L.A., Daniell, H., Depamphilis, C.W., Leebens-Mack, J., Müller, K.F., Guisinger-Bellian, M., Haberle, R.C., Hansen, A.K., Chumley, T.W., Lee, S.B., Peery, R., McNeal, J.R., Kuehl, J.V., and Boore, J.L., Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns, Proc. Natl. Acad. Sci. U. S. A., 2007, vol. 104, no. 49, pp. 19369–19374.
Odintsova, M.S. and Yurina, N.P., Chloroplast genomics of land plants and algae, in Biotechnological Applications of Photosynthetic Protein: Biochips, Biosensors and Biodevices, US: Springer, 2006, pp. 57–72.
Chen, S.L. and Phillips, S.M., Orinus (Poaceae), in Flora of China, Wu, Z.Y. and Raven, P.H., Eds., Beijing: Science Press; St. Louis: Missouri Botanical Garden Press, 2006, vol. 22, pp. 464–465.
Su, X., Wu, G.L., Li, L.L., and Liu, J.Q., Species delimitation in plants using the Qinghai–Tibetan Plateau endemic Orinus (Poaceae: Tridentinae) as an example, Ann. Bot., 2015, vol. 116, no. 1, pp. 35–48.
Su, X., Liu, Y.P., Wu, G.L., Luo, W.C., and Liu, J.Q., A taxonomic revision of Orinus (Poaceae) with a new species, O. intermedius, from the Qinghai–Tibet Plateau, Novon, 2017, vol. 25, no. 2, pp. 206–213.
Su, X., Yue, W., and Liu, J.Q., Germplasm collection and preservation of Orinus (Poaceae) in the Qinghai–Tibet Plateau, Plant Diver.Res., 2013, vol. 35, no. 3, pp. 343–347.
Li, H.X., Zhang, D.P., Hao, Y.Q., and Zhao, H.X., Analysis of microbial diversity of root microecosystem of Orinus kokonorica,J. Shenyang Normal Univ: Nat. Sci. Ed., 2016, vol. 34, no. 2, pp. 228–233.
Doyle, J.J. and Doyle, J.L., Isolation of plant DNA from fresh tissue, Focus, 1990, vol. 12, no. 1, pp. 13–15.
Bolger, A.M., Lohse, M., and Usadel, B., Trimmomatic: a flexible trimmer for Illumina sequence data, Bioinformatics, 2014, vol. 30, no. 15, pp. 2114–2120.
Zerbino, D.R. and Birney, E., Velvet: algorithms for de novoshort read assembly using de Bruijn graphs, Genome Res., 2008, vol. 18, no. 5, pp. 821–829.
Lohse, M., Drechsel, O., Kahlau, S., and Bock, R., Organellar Genome DRAW–a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets, Nucleic Acids Res., 2013, vol. 41, pp. W575–W581.
Katoh, K. and Standley, D.M., . MAFFT multiple sequence alignment software version 7: improvements in performance and usability, Mol. Biol. Evol., 2013, vol. 30, no. 4, pp. 772–780.
Swofford, D.L., PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods), version 4, Sunderland, MA: Sinauer Associates. 2002.
Stamatakis, A., RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies, Bioinformatics. 2014, vol. 30, no. 9, pp. 1312–1313.
Yang, M., Zhang, X.W., Liu, G.M., Yin, Y.X., Chen, K.F., Yun, Q.Z., Zhao, D.J., Al-Mssallem, I.S., and Yu, J., The complete chloroplast genome sequence of date palm (Phoenix dactylifera L.), PLoS One, 2010, vol. 5, no. 9. e12762.
Liu, Y.P., Su, X., Lu, T., Liu, T., and Chen, K.L., Characterization of the complete chloroplast genome sequence of Littledalea racemosa Keng (Poaceae: Bromeae), Conserv. Genet. Resour., 2018, vol. 10, no. 3, pp. 343–346.
Su, X., Liu, Y.P., Lu, T., Liu, T., and Zhu, D., Complete chloroplast genome of Psammochloa villosa (Poaceae), a pioneer grass endemic to sand dunes in northwest China, Conserv. Genet. Resour., 2017, vol. 10.
ACKNOWLEDGMENTS
The authors thank anonymous reviewers for their comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
FUNDING
This work was financially supported by the National Natural Science Foundation of China (41761009, 31260052), the Natural Science Foundation of Qinghai Province (2017-ZJ-904), the State Scholarship Fund of China Scholarship Council (201508635003) and the Young and Middle-Aged Research Foundation of Qinghai Normal University (2017-33).
COMPLIANCE WITH ETHICAL STANDARDS
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
About this article
Cite this article
Yuping Liu, Lv, T., Liu, T. et al. Characterization and Phylogenetic Analysis of the Complete Chloroplast Genome of Orinus kokonoricus (Poaceae), an Endemic Species from the Qinghai-Tibet Plateau. Cytol. Genet. 53, 510–514 (2019). https://doi.org/10.3103/S0095452719060045
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.3103/S0095452719060045