Abstract
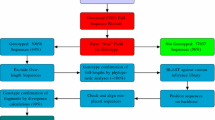
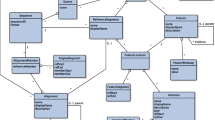
Abstract: To date, more than 30 000 hepatitis C virus (HCV) sequences have been deposited in the generalist databases DNA Data Bank of Japan (DDBJ), EMBL Nucleotide Sequence Database (EMBL) and GenBank®. The main difficulties with HCV sequences in these databases are their retrieval, annotation and analyses. To help HCV researchers face the increasing needs of HCV sequence analyses, we developed a specialised database of computer-annotated HCV sequences, called HCVDB. HCVDB is re-built every month from an up-to-date EMBL database by an automated process. HCVDB provides key data about the HCV sequences (e.g. genotype, genomic region, protein names and functions, known 3-dimensional structures) and ensures consistency of the annotations, which enables reliable keyword queries. The database is highly integrated with sequence and structure analysis tools and the SRS (LION bioscience) keywords query system. Thus, any user can extract subsets of sequences matching particular criteria or enter their own sequences and analyse them with various bioinformatics programs available on the same server.
Availability: HCVDB is available from http://hepatitis.ibcp.fr



Similar content being viewed by others
References
Lindenbach BD, Rice CM. Flaviviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields virology. Philadelphia (PA): Lippincott Williams & Wilkins, 2001: 991–1042
Robertson B, Myers G, Howard C, et al. Classification, nomenclature, and database development for hepatitis C virus (HCV) and related viruses: proposals for standardization. International Committee on Virus Taxonomy. Arch Virol 1998; 143: 2493–503
Pawlotsky JM. Hepatitis C virus genetic variability: pathogenic and clinical implications. Clin Liver Dis 2003; 7: 45–66
Kellam P, Mar Albà M. Virus bioinformatics: databases and recent applications. Appl Bioinformatics 2002; 1: 37–42
Stoesser G, Baker W, van den Broek A, et al. The EMBL Nucleotide Sequence Database: major new developments. Nucleic Acids Res 2003; 31: 17–22
Tokita H, Okamoto H, Iizuka H, et al. The entire nucleotide sequences of three hepatitis C virus isolates in genetic groups 7–9 and comparison with those in the other eight genetic groups. J Gen Virol 1998; 79: 1847–57
Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A 1988; 85: 2444–8
Combet C, Blanchet C, Geourjon C, et al. NPS@: network protein sequence analysis. Trends Biochem Sci 2000; 25: 147–50
Combet C, Jambon M, Deleage G, et al. Geno3D: automatic comparative molecular modelling of protein. Bioinformatics 2002; 18: 213–4
Acknowledgements
This work was supported by Reséau National Hépatites of ‘Programme national de recherche fondamentale en microbiologie et maladies infectieuses et parasitaires du Ministére de la Recherche’ and the EU contract QLK2-CT-2002-01329. C. Combet received a doctoral fellowship from Agence National de Recherches sur le Sida (ANRS). The authors gratefully acknowledge Christian Bréchot, Jean Dubuisson, Geneviève Inchauspé and Jean-Michel Pawlotsky for stimulating discussions and their support for HCVDB creation. The authors have no conflicts of interest that are directly relevant to the content of this article.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Combet, C., Penin, F., Geourjon, C. et al. HCVDB. Appl-Bioinformatics 3, 237–240 (2004). https://doi.org/10.2165/00822942-200403040-00005
Published:
Issue Date:
DOI: https://doi.org/10.2165/00822942-200403040-00005