Abstract
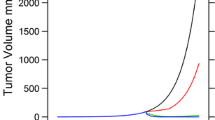
The purpose of this study was to explore the impact of censoring due to animal sacrifice on parameter estimates and tumor volume calculated from two diameters in larger tumors during tumor growth experiments in preclinical studies. The type of measurement error that can be expected was also investigated. Different scenarios were challenged using the stochastic simulation and estimation process. One thousand datasets were simulated under the design of a typical tumor growth study in xenografted mice, and then, eight approaches were used for parameter estimation with the simulated datasets. The distribution of estimates and simulation-based diagnostics were computed for comparison. The different approaches were robust regarding the choice of residual error and gave equivalent results. However, by not considering missing data induced by sacrificing the animal, parameter estimates were biased and led to false inferences in terms of compound potency; the threshold concentration for tumor eradication when ignoring censoring was 581 ng.ml−1, but the true value was 240 ng.ml−1.




Similar content being viewed by others
References
Rubin DB. Inference and missing data. Biometrika. 1976;63(3):581–90. doi:10.1093/biomet/63.3.581.
Little R, Rubin DB. Statistical analysis with missing data, 2nd Edition. John Wiley and Sons ed: John Wiley and Sons; 2002.
Beal SL. Ways to fit a PK model with some data below the quantification limit. J Pharmacokinet Pharmacodyn. 2001;28(5):481–504.
Sausville EA, Burger AM. Contributions of human tumor xenografts to anticancer drug development. Cancer Res. 2006. doi:10.1158/0008-5472.CAN-05-3627.
Rocchetti M, Simeoni M, Pesenti E, De Nicolao G, Poggesi I. Predicting the active doses in humans from animal studies: a novel approach in oncology. Eur J Cancer. 2007;43(12):1862–8. doi:10.1016/j.ejca.2007.05.011.
Pierrillas PB, Tod M, Amiel M, Chenel M, Henin E. Improvement of parameter estimations in tumor growth inhibition models on xenografted animals: a novel method to handle the interval censoring caused by measurement of smaller tumors. AAPS J. 2016. doi:10.1208/s12248-015-9862-1.
Bergstrand M, Karlsson MO. Handling data below the limit of quantification in mixed effect models. AAPS J. 2009;11(2):371–80. doi:10.1208/s12248-009-9112-5.
Sheiner LB, Grasela TH. An introduction to mixed effect modeling: concepts, definitions, and justification. Journal of Pharmacokinetics and Biopharmaceutics. 1991;19.
Lindbom L, Ribbing J, Jonsson EN. Perl-speaks-NONMEM (PsN)—a Perl module for NONMEM related programming. Comput Methods Prog Biomed. 2004;75(2):85–94. doi:10.1016/j.cmpb.2003.11.003.
Lindbom L, Pihlgren P, Jonsson EN. PsN-toolkit—a collection of computer intensive statistical methods for non-linear mixed effect modeling using NONMEM. Comput Methods Prog Biomed. 2005;79(3):241–57. doi:10.1016/j.cmpb.2005.04.005.
Beal SL, Sheiner LB, Boeckmann A, Bauer R. NONMEM user’s guide (1989–2009). Ellicott City: Icon Development Solutions; 2009.
Koch G, Walz A, Lahu G, Schropp J. Modeling of tumor growth and anticancer effects of combination therapy. J Pharmacokinet Pharmacodyn. 2009;36(2):179–97. doi:10.1007/s10928-009-9117-9.
Wang Y. Derivation of various NONMEM estimation methods. J Pharmacokinet Pharmacodyn. 2007;34(5):575–93. doi:10.1007/s10928-007-9060-6.
Karlsson MOH, N. A tutorial on visual predictive checks. Population Approach Group in Europe (2008). Marseille (France) 2008.
Holford N. The visual predictive check—superiority to standard diagnostic (Rorschach) plots. Population Approach Group in Europe (2005). Pamplona (Spain) 2005.
Simeoni M, Magni P, Cammia C, De Nicolao G, Croci V, Pesenti E, et al. Predictive pharmacokinetic-pharmacodynamic modeling of tumor growth kinetics in xenograft models after administration of anticancer agents. Cancer Res. 2004;64(3):1094–101.
Simeoni M, Poggesi I, Germani M, De Nicolao G, Rocchetti M. Population modeling of tumor growth inhibition in vivo: application to anticancer drug development. Population Approach Group in Europe (2004). Uppsala (Sweden)2004.
Sostelly A, Payen L, Guitton J, Di Pietro A, Falson P, Honorat M, et al. Quantitative evaluation of the combination between cytotoxic drug and efflux transporter inhibitors based on a tumour growth inhibition model. Fundam Clin Pharmacol. 2014;28(2):161–9. doi:10.1111/fcp.12005.
Magni P, Simeoni M, Poggesi I, Rocchetti M, De Nicolao G. A mathematical model to study the effects of drugs administration on tumor growth dynamics. Math Biosci. 2006;200(2):127–51. doi:10.1016/j.mbs.2005.12.028.
Ahn JE, Karlsson MO, Dunne A, Ludden TM. Likelihood based approaches to handling data below the quantification limit using NONMEM VI. J Pharmacokinet Pharmacodyn. 2008;35(4):401–21. doi:10.1007/s10928-008-9094-4.
Parra-Guillen ZP, Berraondo P, Grenier E, Ribba B, Troconiz IF. Mathematical model approach to describe tumour response in mice after vaccine administration and its applicability to immune-stimulatory cytokine-based strategies. AAPS J. 2013;15(3):797–807. doi:10.1208/s12248-013-9483-5.
Parra-Guillen ZP, Berraondo P, Ribba B, Troconiz IF. Modeling tumor response after combined administration of different immune-stimulatory agents. J Pharmacol Exp Ther. 2013;346(3):432–42. doi:10.1124/jpet.113.206961.
Martin E. Models to account for dropouts in xenograft experiments due to tumour burden limit. Chester: PKUK; 2015.
Laajala TD, Corander J, Saarinen NM, Makela K, Savolainen S, Suominen MI, et al. Improved statistical modeling of tumor growth and treatment effect in preclinical animal studies with highly heterogeneous responses in vivo. Clinical Cancer Res Off J Am Assoc Cancer Res. 2012;18(16):4385–96. doi:10.1158/1078-0432.CCR-11-3215.
Tan M, Fang HB, Tian GL, Houghton PJ. Small-sample inference for incomplete longitudinal data with truncation and censoring in tumor xenograft models. Biometrics. 2002;58(3):612–20.
Tan M, Fang HB, Tian GL, Houghton PJ. Repeated-measures models with constrained parameters for incomplete data in tumour xenograft experiments. Stat Med. 2005;24(1):109–19. doi:10.1002/sim.1775.
Zhao L, Morgan MA, Parsels LA, Maybaum J, Lawrence TS, Normolle D. Bayesian hierarchical changepoint methods in modeling the tumor growth profiles in xenograft experiments. Clin Cancer Res Off J Am Assoc Cancer Res. 2011;17(5):1057–64. doi:10.1158/1078-0432.CCR-10-1935.
Ayers GD, McKinley ET, Zhao P, Fritz JM, Metry RE, Deal BC, et al. Volume of preclinical xenograft tumors is more accurately assessed by ultrasound imaging than manual caliper measurements. J Ultrasound Med. 2010;29(6):891–901.
Acknowledgments
Emilie Hénin was funded by Fondation Synergie Lyon Cancer and La Ligue Nationale contre le Cancer. This work is part of a PhD project (Philippe Pierrillas) granted by Servier Laboratories. We want to thank François Bouzom for his helpful advice and suggestions. We also want to thank Jacques Pradel for his advice on missing data.
Author information
Authors and Affiliations
Corresponding author
ELECTRONIC SUPPLEMENTARY MATERIAL
Below is the link to the electronic supplementary material.
ESM 1
(PDF 900 kb)
Appendices
Appendix 1
Determination of the Uncertainty on Tumor Size Issued from Xenograft Experiments
The tumor size calculation can be performed using the formula
where p and q are the diameters (length and width) of the ovoid.
If we consider ε the error made on a diameter measure, we have
which leads after development to
After removing the high-order negligible terms, we obtain
Regarding the units of the expression, \( \frac{p\cdot {q}^2}{2} \) is in a volume unit (mm3) and p · q and q 2 have the dimension of an area (mm2) which corresponds to a volume raised to a 2/3 power.
Thereby, we have
with g representing a function of the volume.
Appendix 2
NONMEM Control Stream for the M3-Like Method Considering the Sacrifice Censoring
$PROBLEM SSE Tumor Growth
$DATA
$INPUT
$SUBROUTINE ADVAN13 TOL = 6
$MODEL NCOMP = 3
COMP = DEP; Depot compartment 1
COMP = CENT; Central compartment 2
COMP = TUMOR_GR; Tumor growth 3
$PK
F1 = 1
KA = 1
K10 = 0.1386
V2 = 5
S2 = V2
LAMB1 = THETA(1)*EXP(ETA(1))
LAMB2 = THETA(2)*EXP(ETA(2))
TG0 = THETA(3)*EXP(ETA(3))
K2 = THETA(4)*EXP(ETA(4))
A_0(1) = 0; Initialization of depot compartment
A_0(2) = 0; Initialization of central compartment
A_0(3) = TG0; Initialization of tumor growth compartment
$DES
;------------------------------------------------------PK model------------------------;
CP = A(2)/S2
DADT(1) = −KA*A(1); Depot compartment first-order absorption
DADT(2) = KA*A(1) − K10*A(2); Central compartment first-order elimination
;-----------------------------------------------Tumor growth model-------------------------;
DADT(3) = ((2*LAMB1*LAMB2*A(3))/((LAMB2 + 2*LAMB1*A(3)))) − K2*CP*A(3)
;Biphasic Koch tumor growth with linear antitumor effect
$ERROR
;----------------------------------------------Residual error model-------------------------;
IPRED = A(3)
W = ((THETA(5)**2)*(IPRED**2) + (THETA(6)**2))**(0.5); Weighting of the combined residual error model
IF (DV.LT.2500) THEN; values below 2500 mm3 treated as continuous data
F_FLAG = 0
Y = IPRED + W*EPS(1)
ENDIF
ULOQ = 2500
IF (DV.GE.ULOQ) THEN; Values above 2500 mm3 treated as categorical data
F_FLAG = 1
DUM = (IPRED-ULOQ)/W
PROB = PHI(DUM); Definition of the probability tumor volume to be >2500 mm3
Y = PROB
ENDIF
IRES = DV-IPRED; Definition of individual residuals
IWRES = IRES/W; Definition of individual weighted residuals
$THETA; Initial estimates for typical values
$SIGMA
1 FIX; EPS 1
$OMEGA; Initial estimates for random effects
$ESTIMATION METHOD = 1 INTER NOABORT MAXEVAL = 9999 LAPLACIAN NUMERICAL SLOW;; Estimation with Laplacian options
$TABLE;; Output tables
Rights and permissions
About this article
Cite this article
Pierrillas, P.B., Tod, M., Amiel, M. et al. Improvement of Parameter Estimations in Tumor Growth Inhibition Models on Xenografted Animals: Handling Sacrifice Censoring and Error Caused by Experimental Measurement on Larger Tumor Sizes. AAPS J 18, 1262–1272 (2016). https://doi.org/10.1208/s12248-016-9936-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1208/s12248-016-9936-8