Abstract
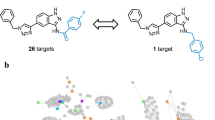
Given the increasing notion of target promiscuity of bioactive compounds and polypharmacological drug behavior, a detailed analysis of publicly available compound activity data from medicinal chemistry sources was carried out to determine and quantify the degree of promiscuity of active compounds across all known human target families. The results are surprising. Approximately 62% of currently available compounds with high-confidence activity data are only annotated with a single biological target, whereas 36% are known to act against multiple targets within the same family (i.e., closely related targets). However, only ∼2% of bioactive compounds are promiscuous across different target families. Thus, despite general data sparseness, these findings indicate that highly promiscuous bioactive compounds only rarely occur. Because pharmaceutically relevant active compounds represent the pool from which drug candidates emerge, one might extrapolate from these results and conclude that there is a low statistical probability to obtain drugs that act against multiple targets belonging to distinct families.






Similar content being viewed by others
REFERENCES
Paolini GV, Shapland RHB, van Hoorn WP, Mason JS, Hopkins AL. Global mapping of pharmacological space. Nat Biotechnol. 2006;24(7):805–15. doi:10.1038/nbt1228.
Keiser MJ, Roth BL, Armbruster BN, Ernsberger P, Irwin JJ, Shoichet BK. Relating protein pharmacology by ligand chemistry. Nat Biotechnol. 2007;25(2):197–206. doi:10.1038/nbt1284.
Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4(11):682–90. doi:10.1038/nchembio.118.
Keiser MJ, Setola V, Laggner C, Abbas AI, Hufeisen SJ, Jensen NH, Kuijer MB, Matos RC, Tran TB, Whaley R, Glennon RA, Hert J, Thomas KL, Edwards DD, Shoichet BK, Roth BL. Predicting new molecular targets for known drugs. Nature. 2009;462(7270):175–81. doi:10.1038/nature08506.
Mestres J, Gregori-Puigjané E. Conciliating binding efficiency and polypharmacology. Trends Pharmacol Sci. 2009;30(9):470–4. doi:10.1016/j.tips.2009.07.004.
Merino A, Bronowska AK, Jackson DB, Cahill DJ. Drug profiling: knowing where it hits. Drug Discov Today. 2010;15(17–18):749–56. doi:10.1016/j.drudis.2010.06.006.
Mestres J, Gregori-Puigjane E, Valverde S, Sole RV. Data completeness—the Achilles heel of drug-target networks. Nat Biotechnol. 2008;26(9):983–4. doi:10.1038/nbt0908-983.
Koutsoukas A, Simms B, Kirchmair J, Bond PJ, Whitmore AV, Zimmer S, Young MP, Jenkins JL, Glick M, Glen RC, Bender A. From in silico target prediction to multi-target drug design: current databases, methods and applications. J Proteomics. 2011;74(12):2554–74. doi:10.1016/j.jprot.2011.05.011.
Xie L, Xie L, Kinnings SL, Bourne PE. Novel computational approaches to polypharmacology as a means to define responses to individual drugs. Annu Rev Pharmacol Toxicol. 2012;52:361–79. doi:10.1146/annurev-pharmtox-010611-134630.
Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov. 2004;3(8):673–83. doi:10.1038/nrd1468.
Chong CR, Sullivan DJ. New uses for old drugs. Nature. 2007;448(7154):645–6. doi:10.1038/448645a.
Hu Y, Bajorath J. Polypharmacology directed compound data mining: identification of promiscuous chemotypes with different activity profiles and comparison to approved drugs. J Chem Inf Model. 2010;50(12):2112–8. doi:10.1021/ci1003637.
Bender A, Scheiber J, Glick M, Davies JW, Azzaoui K, Hamon J, Urban L, Whitebread S, Jenkins JL. Analysis of pharmacology data and the prediction of adverse drug reactions and off-target effects from chemical structure. ChemMedChem. 2007;2(6):861–73. doi:10.1002/cmdc.200700026.
Brown JB, Okuno Y. Systems biology and systems chemistry: new directions for drug discovery. Chem Biol. 2012;19(1):23–8. doi:10.1016/j.chembiol.2011.12.012.
Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A, Light Y, McGlinchey S, Michalovich D, Al-Lazikani B, Overington JP. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012;40(D1):D1100–7. doi:10.1093/nar/gkr777.
UniProtConsortium. Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res. 2012;40(D1):D142–8. doi:10.1093/nar/gkr981.
Bemis GW, Murcko MA. The properties of known drugs. 1. Molecular frameworks. J Med Chem. 1996;39(15):2887–93. doi:10.1021/jm9602928.
Molecular Operating Environment (MOE), 2011.10; Chemical Computing Group Inc., 1010 Sherbooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7, 2011.
Knox C, Law V, Jewison T, Liu P, Ly S, Frolkis A, Pon A, Banco K, Mak C, Neveu V, Djoumbou Y, Eisner R, Guo AC, Wishart DS. DrugBank 3.0: a comprehensive resource for ‘omics’ research on drugs. Nucleic Acids Res. 2011;39(Database issue):D1035–41. doi:10.1093/nar/gkq1126.
Hu Y, Bajorath J. Many structurally related drugs bind different targets whereas distinct drugs display significant target overlap. RSC Adv. 2012;2(8):3481–9. doi:10.1039/C2RA01345B.
Hopkins AL, Mason JS, Overington JP. Can we rationally design promiscuous drugs? Curr Opin Struct Biol. 2006;16(1):127–36. doi:10.1016/j.sbi.2006.01.013.
Fabian MA, Biggs III WH, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, Carter TA, Ciceri P, Edeen PT, Floyd M, Ford JM, Galvin M, Gerlach JL, Grotzfeld RM, Herrgard S, Insko DE, Insko MA, Lai AG, Lélias JM, Mehta SA, Milanov ZV, Velasco AM, Wodicka LM, Patel HK, Zarrinkar PP, Lockhart DJ. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol. 2005;23(3):329–36. doi:10.1038/nbt1068.
Sturm N, Desaphy J, Quinn RJ, Rognan D, Kellenberger E. Structural insights into the molecular basis of the ligand promiscuity. J Chem Inf Model. 2012;52(9):2410–21. doi:10.1021/ci300196g.
Evans BE, Rittle KE, Bock MG, Dipardo RM, Freidinger RM, Whitter WL, Lundell GF, Veber DF, Anderson PS. Methods for drug discovery: development of potent, selective, orally effective cholecystokinin antagonists. J Med Chem. 1988;31(12):2235–46. doi:10.1021/jm00120a002.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
Figure S1 (activity profiles) and Tables S1–S11 (activity profiles, target families, and compound statistics). (DOC 364 kb)
Rights and permissions
About this article
Cite this article
Hu, Y., Bajorath, J. How Promiscuous Are Pharmaceutically Relevant Compounds? A Data-Driven Assessment. AAPS J 15, 104–111 (2013). https://doi.org/10.1208/s12248-012-9421-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1208/s12248-012-9421-y