Abstract
The NAC (NAM, ATAF and CUC) family is one of the largest plant-specific transcription factor (TF) families. Members of this family are implicated in plant growth, development and stress responses. Recent functional studies demonstrate that a number of NAC TFs function as positive or negative regulators of plant immunity to biotrophic, hemibiotrophic or necotrophic pathogens, as modulators of the hypersensitive responses and stomatal immunity or as virulence targets of pathogen effectors. They affect plant immunity through their regulatory impact on signaling of plant hormones, which in turn are key players in plant immune responses. This review summarizes current knowledge and recent progress in our understanding of the biological functions of NAC TFs in plant immunity and discusses perspectives and directions for further study to elucidate the molecular mechanisms of NAC TF functions in immunity and potential application in improvement of crop disease resistance.
Similar content being viewed by others
Background
To combat pathogen attack, plants have evolved sophisticated immunity systems. Two distinct types of innate immune responses, known as pathogen/microbe/damage-associated molecular pattern (PAMP/MAMP/DAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI), have been recognized in plants (Jones and Dangl 2006; Boller and He 2009; Spoel and Dong 2012). PTI forms the first layer of immunity toward all potential microbial attack, whereas ETI is a specialized form of immunity triggered by the direct or indirect interactions between plant R proteins and pathogen effector proteins (Zhang and Zhou 2010; Segonzac and Zipfel 2011; Schwessinger and Ronald 2012; Bigeard et al. 2015; Cui et al. 2015). Upon perception of pathogen-derived signals, plants often activate a network of defense signaling pathways (Pieterse et al. 2009; Peng et al. 2018), which ultimately lead to transcriptional reprogramming that coordinately regulates expression of a large set of genes (Tsuda and Somssich 2015; Li et al. 2016). Such large-scale transcriptional reprogramming of gene expression in a specific immune response obviously requires a concerted function of different types of transcription factors (TFs) in both temporal and spatial manners (Buscaill and Rivas 2014; Birkenbihl et al. 2017). Recent genetic studies have demonstrated that a number of TFs from the families of WRKY, AP2/ERF (Apetala2/Ethylene Responsive Factor), NAC (NAM, ATAF and CUC), MYB, bZIP (Basic leucine zipper domain), bHLH (Basic helix-loop-helix), NF-Y (Nuclear Factor Y) and CAMTA (CaM-binding transcription activator) play crucial roles in immune responses against pathogens (Nuruzzaman et al. 2013; Buscaill and Rivas 2014; Huang et al. 2016; Phukan et al. 2016; Noman et al. 2017; Zanetti et al. 2017).
The NAC proteins constitute a large plant-specific TFs family, which contains more than 100 members in Arabidopsis and rice (Ooka et al. 2003; Fang et al. 2008; Nuruzzaman et al. 2010). NAC TFs are characterized by the presence of a highly conserved N-terminal region, known as NAC domain, which functions as a DNA-binding domain and is also responsible for oligomerization into dimer (Olsen et al. 2005; Puranik et al. 2012). The C-terminal region of NAC TFs is more diverse, intrinsically disordered and functions as a transcriptional regulatory domain (Olsen et al. 2005; Jensen et al. 2010). On the structural basis, the NAC TFs can be divided into two classes, typical NAC TFs and atypical NAC TFs. Whereas the typical NAC TFs contain a conserved N-terminal NAC domain and a divergent C-terminal region (Olsen et al. 2005; Puranik et al. 2012), the atypical NAC proteins are characterized, in addition to the NAC domain, by the presence of additional conserved domains/motifs in the C-terminal regions or the absence of the C-terminus (Puranik et al. 2012). One type of atypical NAC TFs is the NTL (NAC with transmembrane motif1-like) proteins, which are featured by the presence of a variable transcriptional regulatory domain and a transmembrane (TM) motif in the C-terminal region (Ernst et al. 2004). The TM motif in NTLs is thought to be responsible for anchoring to plasma membrane or endoplasmic reticulum membrane where they could be released by proteolysis to exert their function (Kim et al. 2007a; Liang et al. 2015).
During the last two decades, extensive functional studies have shown that, in addition to their functions in plant growth, development and abiotic stress responses (Zhong et al. 2010; Bollhoner et al. 2012; Nakashima et al. 2012; Puranik et al. 2012; Hussey et al. 2013; Nuruzzaman et al. 2013; Ricachenevsky et al. 2013; Shao et al. 2015; Kim et al. 2016), a large number of the NAC TFs play critical roles in plant immunity. In this review, we summarize the biological functions of the NAC TFs in plant immunity against pathogens and discuss further directions to investigate the molecular mechanisms of NAC TFs in immunity as well as the perspectives to improve crop disease resistance using NAC TFs as targets.
Functions of NAC TFs in plant immunity
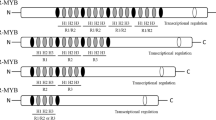
Plant pathogens are classified as biotrophs, hemibiotrophs and necrotrophs according to their lifestyles (Wang et al. 2014). The biotrophs generally feed on living hosts while the hemibiotrophs possess a biotrophic phase during the early stage of infection. In contrast, necrotrophs can actively kill hosts to acquire nutrients (Lai and Mengiste 2013). Functional analyses of NAC TFs in plant immunity have been extensively performed using knockout/knockdown mutants and overexpression lines in Arabidopsis, rice and other plant species. Dozens of NAC genes have been demonstrated to play important roles in plant immunity by acting as negative or positive regulators, modulator of hypersensitive response and stomatal immunity or targets of pathogen effectors (Table 1). These reported immunity-related NAC TFs belong to different subfamilies of the NAC family (Fig. 1).
Phylogenetic tree of the reported immunity-related NAC TFs. The tree was constructed using the Neighbour-Joining algorithm method and bootstrap values from 1000 replicates are indicated at each node. The GenBank accession numbers are as follows: OsNAC6, BAA89800; HvNAC6, AM500854; ATAF1, NP_171677.1; ATAF2, NP_680161; GhATAF1, DT549350.1; AaNAC1, AQU15092.1; ANAC072/RD26, NP_001078452.1; ANAC019, NP_175697; ANAC055, NP_188169.1; JA2, NP_001233972.1; JA2L, NP_001306107.1; TaNAC1, AEI00649.1; VvNAC1, XP_002282566; SlNAC35, XM_004236996; ONAC122, BAF27472.1; ONAC131, BAT15671.1; ANAC042/JUB1, NP_181828; NAC4, NP_568182.2; TaNAC21/22, AGV08300.1; ANAC062/NTL6, NP_190522.1; StNTP1, AGY49284.1; CBNAC/NTL9, NP_001119122.1; RIM1, BAS82017.1; SlSRN1, NP_001304297.1; StNTP2, AGY49285.1; VpNAC1, GU393316; OsNAC111, XP_015615861.1; GbNAC1, AOW72925.1
Functions of NAC TFs in plant immunity against biotrophic and hemibiotrophic pathogens
Several NAC TFs have been demonstrated to play roles in different stages of plant immunity against biotrophic and hemibitrophic pathogens. In the non-host Blumeria graminis f. sp. hordei (Bgh)-Arabidopsis interaction, ATAF1, belonging to the ATAF subfamily (Fig. 1), promotes penetration resistance as the ataf1–1 mutant plants showed decreased penetration resistance to Bgh (Jensen et al. 2007). Similarly, silencing of HvNAC6, the ATAF1 homologue in barley (Fig. 1), compromised penetration resistance in epidermal cells towards virulent Bgh while transient overexpression of HvNAC6 increased the occurrence of penetration resistant cells towards Bgh attack (Jensen et al. 2007). Thus, it was proposed that ATAF1 and its homologues play a conserved role in penetration resistance in dicot and monocot plants. Overexpression of ATAF2, closely related to ATAF1 in the ATAF subfamily (Fig. 1), led to a significant reduction in virus accumulation, accompanied with upregulated expression of defense genes PR1, PR2 and PDF1.2, indicating that ATAF2 functions in the regulation of host basal defense (Wang et al. 2009b). Overexpression of ATAF1 resulted in increased susceptibility while suppression of ATAF1 led to enhanced resistance in transgenic Arabidopsis plants to Pseudomonas syringae pv. tomato (Pst) DC3000 (Wang et al. 2009a; Wu et al. 2009). The ANAC019, ANAC055 and ANAC072 are three members of the NAM subfamily (Fig. 1) and the anac019 anac055 anac072 triple mutant plants exhibited enhanced resistance to P. syringae pv. maculicola ES4326 (Bu et al. 2008; Zheng et al. 2012). Overexpression of ANAC042/JUB1, a reactive oxygen species-responsive NAC gene belonging to NAM subfamily (Fig. 1), attenuated Arabidopsis resistance to Pst DC3000, while the anac042/jub1 knockdown mutant exhibited reduced disease symptoms and growth of the bacterial pathogen Pst DC3000, indicating that ANAC042/JUB1 suppresses Arabidopsis immunity against Pst DC3000 (Wu et al. 2012; Shahnejat-Bushehri et al. 2016a, b).
The Arabidopsis NCBAC/NTL9, a member of the NTL subfamily (Fig. 1), plays distinct roles in different stages of plant immunity. The cbnac/ntl9 mutant plants displayed enhanced resistance to a virulent strain of Pst DC3000 while resistance was reduced in CBNAC/NTL9-overexpressing plants, and the changes in resistance were correlated with changes in defense gene expression (Kim et al. 2012; Guo et al. 2017). By contrast, the ncbac1/ntl9 mutant plants displayed reduced resistance to P. syringae hopD1 mutant and ETI-inducing P. syringae strains and failed to induce the expression of SA biosynthesis genes in response to flg22, whereas transgenic plants expressing a constitutively active CBNAC/NTL9 derivative showed increased resistance to these P. syringae strains (Zheng et al. 2012; Block et al. 2014). These contradictory results may indicate that NCBAC1/NTL9 plays distinct roles at different stages or aspects of plant immunity. NTL6 (ANAC062), another member of the NTL subfamily (Fig. 1), is cold-induced and its transcriptionally active form, 6△C, which was modified by deleting its C-terminal region, entered the nucleus and induced a subset of cold-responsive defense genes PR1, PR2 and PR5. Transgenic plants overexpressing the active NTL6 form, 6△C, exhibited enhanced disease resistance, whereas NTL6-RNAi plants with reduced NTL6 were more susceptible to infection by a virulent strain of Pst DC3000 at low temperatures, indicating that NTL6 integrates cold signals into plant defense responses (Seo et al. 2010).
In rice, transgenic plants overexpressing OsNAC6, the rice ATAF1 homologue (Fig. 1), displayed increased resistance to Magnaporthe grisea (Nakashima et al. 2007), the causal agent of rice blast disease. Silencing of ONAC122 or ONAC131, two closely related rice NAC TFs that belong to the NAP subfamily (Fig. 1), caused increased susceptibility to M. oryzae and downregulated expression of several defense- and signaling-related genes (i.e. OsLOX, OsPR1a, OsWRKY45 and OsNH1), suggesting that both ONAC122 and ONAC131 positively regulate rice blast resistance (Sun et al. 2013). OsNAC111-overexpressing plants showed increased resistance to M. oryzae and constitutively expressed several defense genes, suggesting that OsNAC111, a member of TERN subfamily (Fig. 1), positively regulates the expression of a specific set of defense genes and contributes to disease resistance (Yokotani et al. 2014). Overexpression of OsNAC066, which was induced by the blast fungus, increased resistance to M. oryzae and Xanthomonas oryzae pv. oryzae (Liu et al. 2018). Mutation in OsNAC60 increased susceptibility of the osnac60 plants to M. oryzae (Wang et al. 2018a), while overexpression of OsNAC58 increased resistance to X. oryzae pv. oryzae (Park et al. 2017). Disruption of RIM1, coding for a rice NAC TF belonging to the NTL subfamily (Fig. 1), resulted in reduced rice susceptibility to Rice dwarf virus (RDV) but remains susceptible to other viruses. The accumulation of RDV capsid proteins was drastically reduced in rim1 mutant plants, indicating that the multiplication of RDV is specifically impaired in the rim1 mutant. It was proposed that RIM1 negatively regulates rice immunity to RDV by acting as a host factor that is required for multiplication of RDV in host plants (Yoshii et al. 2009).
Functions of NAC TFs from other plant species have also been explored through virus-induced gene silencing or antisense-mediated suppression in host plants and ectopic expression in Arabidopsis or tobacco. Tomato JA2 and JA2L, two closely related NAC TFs belonging to the AtNAC3 subfamily (Fig. 1), play different roles in regulating immunity against Pst DC3118 by modulating pathogen-induced stomatal closure and reopening (Du et al. 2014). Silencing of SlSRN1 in tomato plants resulted in decreased resistance against Pst DC3000 (Liu et al. 2014), whereas ectopic overexpression of SlNAC35 in tobacco improved resistance to Ralstonia solanacearum, accompanied with increased expression of SA-responsive defense genes PR1a, NPR1, PR2 and PR5 (Wang et al. 2016a). Overexpression of grapevine VvNAC1 and VpNAC1 or Artemisia annua AaNAC1 in Arabidopsis or tobacco plants led to enhanced resistance to Hyaloperonospora arabidopsidis and Erysiphe cichoracearum (Zhu et al. 2012; Le Hénanff et al. 2013; Lv et al. 2016). Knockdown of TaNAC1, TaNAC21/22 or TaNAC30 in wheat enhanced resistance against Puccinia striiformis f. sp. tritici, indicating that TaNAC1, belonging to NAP subfamily (Fig. 1), TaNAC21/22, belonging to NAM subfamily (Fig. 1), and TaNAC30 negatively regulate the stripe rust resistance in wheat (Feng et al. 2014; Wang et al. 2015a; Wang et al. 2018b). By contrast, overexpression of TaNAC6-A enhanced resistance against Blumeria graminis f. sp. tritici (Bgt), while silencing of each TaNAC6s compromised the resistance, suggesting that TaNAC6s play positive roles in broad-spectrum resistance against Bgt (Zhou et al. 2018).
Functions of NAC TFs in plant immunity against necrotrophic pathogens
In Arabidopsis, at least seven NAC proteins ATAF1, ATAF2, ANAC019, ANAC055, ANAC072, NTL9/CBNAC (Calmodulin-binding NAC protein) and ANAC042/JUB1 (JUNGBRUNNEN1) are implicated in the regulation of plant immunity to necrotrophic fungal pathogens including Botrytis cinerea, Alternaria brassicicola, and Fusarium oxysporum. We and others found that transgenic plants overexpressing ATAF1 (ATAF1-OE) displayed increased susceptibility while transgenic plants expressing an ATAF1 chimeric repressor (ATAF1-SRDX) construct enhanced resistance to B. cinerea and A. brassicicola (Wang et al. 2009a; Wu et al. 2009). Expressions of defense genes were upregulated in the ATAF1-SRDX plants but attenuated or unchanged in the ATAF1-OE plants, after inoculation with B. cinerea or Pst DC3000 (Wang et al. 2009a). Similarly, overexpression of GhATAF1, a homologue of ATAF1 in cotton (Fig. 1), decreased resistance to B. cinerea and Verticilium dahlia, coupled with the suppression of JA-mediated signaling and the activation of SA-mediated signaling (He et al. 2016). The anac019 anac055 double mutant plants showed increased resistance to B. cinerea (Bu et al. 2008; Zheng et al. 2012). The JA-induced expression of VSP1 and LOX2 in anac019 anac055 double mutant plants was attenuated while their expression in transgenic plants overexpressing either ANAC019 or ANAC055 was upregulated (Bu et al. 2008). It was found that overexpression of ATAF2 increased susceptibility to Fusarium oxysporum and reduced expression of defense genes in transgenic Arabidopsis plants (Delessert et al. 2005). Furthermore, mutation in ANAC042/JUB1 led to enhanced susceptibility to A. brassiscicola and reduced accumulation of camalexin, indicating that ANAC042/JUB1 regulates immunity against A. brassicicola through modulating the camalexin biosysnthesis in Arabidopsis plants (Saga et al. 2012).
Additionally, silencing of SlSRN1 in tomato plants resulted in increased susceptibility to B. cinerea (Liu et al. 2014), whereas silencing of potato StNTP1 and StNTP2, two membrane-localized NAC TFs belonging to the NTL subfamily (Fig. 1), in Nicotiana benthamiana increased susceptibility to Phytophthora infestans infection (McLellan et al. 2013). GbNAC1-silenced cotton plants attenuated resistance to Verticillium dahlia while GbNAC1-overexpressing Arabidopsis plants enhanced resistance to V. dahliae (Wang et al. 2016b). Overexpression of grapevine VvNAC1 or Artemisia annua AaNAC1 in Arabidopsis or tobacco plants enhanced resistance to B. cinerea or P. parasitica var. nicotianae (Zhu et al. 2012; Le Hénanff et al. 2013).
NACs as modulators of hypersensitive response
Hypersensitive response (HR) is a form of programmed cell death (PCD) and is an active and efficient primary immune response that prevents pathogen invasion in plants (Coll et al. 2011). The function of rice OsNAC4 in HR cell death has been well studied (Kaneda et al. 2009). OsNAC4 was found to be upregulated during non-host defense response and regulate the occurrence of HR cell death. In response to an avirulent strain of Acidovorax avenae, the OsNAC4 knockdown plants decreased while the OsNAC4-overexpressing plants enhanced HR cell death, accompanied by loss of plasma membrane integrity, nuclear DNA fragmentation and typical morphological changes (Kaneda et al. 2009), which are characteristics of PCD (Coll et al. 2011). On the other hand, Arabidopsis NAC4, a member of NAM subfamily (Fig. 1), plays an essential role in the regulation of HR cell death. HR cell death was noticeably enhanced in NAC4-overexpressing plants in response to an avirulent strain of Pst DC3000 (AvrRpm1), demonstrating that NAC4 is a positive regulator of HR cell death (Lee et al. 2017).
NACs as modulators of stomatal immunity
Stomata are not just a passive port for pathogen entry but also play an active role in the plant innate immune response (Arnaud and Hwang 2015). The control of stomatal closure is one of the first lines of defense against pathogen invasion and thus stomatal opening or closing, governed by guard cells, is the main battleground during the early stage of plant-pathogen interaction. Recent discoveries have revealed that some specific NAC TFs such as the Arabidopsis ANAC019/ANAC055/ANAC072 and CBNAC/NTL9 and tomato JA2 and JA2L participate in signaling pathways that regulate stomatal immunity. In the Arabidopsis anac019 anac055 anac72 triple mutant plants, Pst DC3000-induced stomatal closure occurred at 1 hours post inoculation (hpi) but Pst DC3000-induced stomatal reopening did not occur and the stomatal apertures remained small at 4 hpi, suggesting that ANAC019/ANAC055/ANAC072 are required for Pst DC3000-triggered stomatal reopening and thus play critical role in stomatal-mediated immunity. Both JA2 and JA2L, encoding two NAC TFs that are closely related to ANAC019/ANAC055/ANAC072, were preferentially expressed in guard cells of tomato leaves (Du et al. 2014). The Pst DC3000-induced stomatal closure at 1 hpi was impaired and the pathogen-triggered stomatal reopening at 4 hpi remained normal in JA2-SRDX plants, indicating that JA2 is required for Pst DC3000-induced stomatal closure but not stomatal reopening (Du et al. 2014). By contrast, the Pst DC3000-induced stomatal closure at 1 hpi was largely normal but the pathogen-triggered stomatal reopening at 4 hpi was substantially impaired in JA2L-AS plants, indicating that JA2L is required for pathogen-regulated stomatal reopening (Du et al. 2014). It was speculated that JA2 acts in ABA-mediated stomatal closure while JA2L acts in JA-mediated stomatal reopening, showing distinct functions of JA2 and JA2L in stomatal immunity against Pst DC3000. Furthermore, the Arabidopsis CBNAC/NTL9 is preferentially expressed in guard cells (Yoon et al. 2008; Zheng et al. 2015) and the cbnac/ntl9 mutant plants impaired flg22-induced SA biosynthesis in guard cells and Psm ES4326-induced change of stomatal aperture, demonstrating that CBNAC/NTL9 is specifically required for SA synthesis in guard cells and plays an essential role in the flg22-triggered stomatal immunity (Zheng et al. 2015).
NACs as targets of pathogen effectors
During infection process, pathogens often secrete and inject numerous effectors into host plant cells to suppress immune response. Recent studies have indicated that some NAC proteins are targets of pathogen effectors to facilitate infection by inhibiting plant immune responses. In Arabidopsis, CBNAC/NTL9 was found to physically interact with one effector from P. syringae and four effectors from Hyaloperonospora arabidopsidis (Mukhtar et al. 2011). A recent study revealed that the P. syringae effector HopD1, a strong repressor of ETI, interacted with CBNAC/NTL9 in endoplasmic reticulum (ER) and inhibited CBNAC/NTL9-mediated induction of certain defense genes during ETI (Block et al. 2014). The potato StNTP1 and StNTP2 were found to be targets of RxLR effector Pi03192 secreted by P. infestans (McLellan et al. 2013). StNTP1 and StNTP2 were released from the ER membrane, where they are normally localized, in response to treatment with P. infestans culture filtrate (CF) and then translocated and accumulated in the nucleus, where they contribute to potato immunity to inhibit disease progression. By contrast, Pi03192 prevented CF-triggered re-localization of StNTP1 and StNTP2 from the ER into the nucleus. It is likely that, during infection process, P. infestans-secreted Pi03192 interacts with StNTP1 and StNTP2 and prevents the ER-nucleus translocation of these two host NAC TFs to promote disease progression (McLellan et al. 2013).
It has been found that, during viral infection, NAC TFs can be hijacked by interacting with viral proteins to facilitate viral replication or suppress host immunity involved in plant-virus interactions. An earlier study characterized two wheat NAC TFs, GRAB1 (Geminivirus Rep A-binding) and GRAB2, which interacted with Wheat dwarf geminivirus (WDV) RepA protein (Xie et al. 1999). Overexpression of GRAB1 and GRAB2 in cultured wheat cells severely inhibited WDV DNA replication, indicating the involvement of GRAB1 and GRAB2 in viral DNA replication (Xie et al. 1999). Tomato SlNAC1 was found to interact with geminivirus replication enhancer REn from Tomato leaf curl virus (TLCV), and overexpression of SlNAC1 enhanced the accumulation of TLCV DNA (Selth et al. 2005). Geminiviral replication initiator protein Rep from Mungbean yellow mosaic India virus (MYMIV) was found to interact with Arabidopsis AtNAC083, which plays a possible role in MYMIV DNA replication (Suyal et al. 2014).
Arabidopsis ATAF2 interacted with the helicase domain of the Tobacco mosaic virus (TMV) 126−/183 kDa replicase protein. Overexpression of ATAF2 significantly reduced virus accumulation, indicating that the TMV replicase-ATAF2 interaction suppresses basal host defenses to promote systemic virus accumulation (Wang et al. 2009b). The capsid protein (CP) of Turnip crinkle virus (TCV) interacts with an Arabidopsis NAC protein TIP (TCV-interacting protein) and the TCV CP-TIP interaction prevented nuclear localization of TIP (Ren et al. 2005, 2012). Although mutation in TIP resulted in increased replication of TCV and Cucumber mosaic virus (CMV), TIP is required for basal resistance to CMV but not for resistance to TCV (Jeong et al. 2008). Lack of TIP-CP interaction in TCV mutant with a mutated CP caused more severe symptoms, implying that TCV regulates antiviral basal immunity in host plants through TIP-CP interaction (Donze et al. 2014).
NACs in plant immune signaling
Upon pathogen attack, plants timely perceive the pathogen-derived signals and often activate efficient and complicated but fine-tuned network of defense hormone-mediated signaling pathways (Zhang et al. 2017; Peng et al. 2018). Emerging evidence has indicated that some NAC proteins participate in modulating these immune signaling pathways to execute their critical functions in plant immunity.
NACs in SA signaling
Salicylic acid (SA) is a critical signaling molecule and mediates SA signaling pathway that regulate immunity against biotrophs and hemibiotrophs (Glazebrook 2005; Shigenaga and Argueso 2016). Pathogen-induced SA production is largely synthesized via the isochorismate pathway, in which the isochorismate synthase 1 (ICS1) is a critical enzyme responsible for approximately 90% of the SA production (Vlot et al. 2009). Meanwhile, most of the SA produced in plant is converted into SA O-β-glucoside (SAG) by a pathogen-inducible SA glucosyltransferase (SAGT) (Vlot et al. 2009). It was recently found that, in the anac019 anac055 anac072 triple mutant Arabidopsis plants, the basal transcript level of ICS1 was higher, whereas the basal transcript level of SAGT1 was lower than that in the wild-type plants, suggesting that ANAC019/ANAC055/ANAC072 may decrease SA synthesis, but increase SA metabolism through transcriptional repression of ICS1 and transcriptional activation of SAGT1, respectively (Zheng et al. 2012). Further ChIP experiments revealed that ANAC019-GFP was enriched in some NAC core-binding sites in promoters of the ICS1, SAGT1 and SA methyl transferase 1 (BSMT1, coding an enzyme that converts SA to the inactive methyl SA) (Zheng et al. 2012). Thus, it is likely that ANAC019/ANAC055/ANAC072 act as negative regulators of SA accumulation through the induction of SAGT1 and BSMT1 and repression of ICS1 via direct binding to their promoters. The N-terminal fragment CBNAC/NTL91–330 of the Arabidopsis CBNAC/NTL9 not only strongly activated the expression of the reporter LUC gene under the control of ICS1 promoter but also interacted with the promoters of EDS1, PAD4 and PBS3 genes (Zheng et al. 2015), revealing that CBNAC/NTL9 may coordinately regulate these four SA synthesis-related genes and thus modulate SA accumulation in plants. In tomato, the Pst DC3000-induced expression of SAMT1 and SAMT2, coding for SA methyl transferases that convert SA to the inactive methyl salicylate was markedly weakened in the JA2L-AS plants and the GFP-JA2L proteins were enriched in fragments containing NAC core-binding sites in promoters of the SAMT1 and SAMT2 genes in ChIP experiments, indicating that JA2L represses SA accumulation by activating the transcription of two tomato SAMT genes, SAMT1 and SAMT2 (Du et al. 2014).
NACs in JA signaling
Jasmonates (JA) and ethylene (ET) are critical defense signaling molecules, mediating the JA/ET signaling that regulates immunity against necrotrophs and herbivorous insects (Glazebrook 2005; Pieterse et al. 2012; Shigenaga and Argueso 2016; Zhang et al. 2017). In Arabidopsis, JA- and Psm ES4326-induced expression of ANAC019, ANAC055 and ANAC072 was dependent on MYC2, a master regulator of the JA signaling (Kazan and Manners 2013), and ChIP experiments confirmed the interaction of MYC2 with G box elements in promoters of the ANAC019, ANAC055 and ANAC072 genes, indicating that expression of the ANAC019, ANAC055 and ANAC072 genes was activated by MYC2 through direct interaction (Bu et al. 2008; Zheng et al. 2012). In anac019 anac055 double mutant plants, JA-induced expression of VEGETATIVE STORAGE PROTEIN1 (VSP1) and LIPOXYGENASE2 (LOX2) was attenuated, whereas JA-induced VSP1 and LOX2 expression was enhanced in transgenic plants overexpressing the ANAC019 or ANAC055 gene, which implies that ANAC019 and ANAC055 act downstream of MYC2 to regulate JA-signaled defense responses (Bu et al. 2008). In rice, the expression of genes encoding JA biosynthetic enzymes lipoxygenase (LOX), allene oxide synthase 2 (AOS2) and OPDA reductase 7 (OPR7) was up-regulated in the rim1 mutant and a rapid and massive accumulation of endogenous JA was detected in the rim1 mutant after wounding, suggesting that RIM1 may represent a new molecular link in JA signaling (Yoshii et al. 2010). TaNAC1-overexpression in Arabidopsis suppressed the expression levels of resistance-related genes PR1 and PR2 involved in SA signaling and AtWRKY70, which functions as a connection node between the JA- and SA-signaling pathways (Wang et al. 2015a).
NACs in ABA signaling
Abscisic acid (ABA) is a well-known stress hormone, and ABA signaling plays an important role in plant immunity (Lievens et al. 2017). Nine-cis epoxycarotenoid dioxygenases (NCEDs) are rate-limiting enzymes of ABA biosynthesis while CYP707As negatively regulate ABA accumulation by catalyzing ABA metabolism (Lievens et al. 2017). The JA2-SRDX-1 tomato plants contain reduced ABA levels and exhibit reduced expression of NCED1, indicating that JA2 regulates NCED1 expression for ABA biosynthesis in tomato (Du et al. 2014). Further biochemical experiments revealed that JA2 specifically interacted with the NAC core binding sites of the NCED1 promoter and selectively activated the expression of NCED1, confirming regulation of ABA biosynthesis by JA2 through direct binding to the promoter of NCED1 (Du et al. 2014). In rice, overexpression of ONAC066 remarkably suppresses the expression of ABA-related genes and decreases endogenous ABA level, but does not affect SA- and JA-related genes as well as the endogenous SA and JA levels (Liu et al. 2018). Y1H assays demonstrate that ONAC066 directly binds to the promoters of LIP9 and NCED4 to modulate their expression (Liu et al. 2018). It was also found that HvNAC6-RNAi plants display an altered light/dark rhythm of ABA level, compromised expression of the two ABA biosynthetic genes HvNCED1 and HvNCED2, elevated expression of an ABA conjugating enzyme gene HvBG7, and impaired basal resistance against Bgh by exogenous application of ABA. These observations, suggest that HvNAC6 regulates ABA signaling by modulating circadian control of ABA level (Chen et al. 2013). Comparative transcriptome analyses revealed that an ABA biosynthesis gene ABSCISIC ALDEHYDE OXIDASE3 and a number of ABA-responsive genes were significantly induced by Bgh in ataf1 plants and that Bgh induced reduction in endogenous ABA content was dependent on ATAF1, indicating that ATAF1 attenuates ABA signaling for efficient penetration resistance in Arabidopsis against Bgh (Jensen et al. 2008). However, it was observed that ANAC019/ANAC055/ANAC072-mediated stomatal immunity against Psm ES4326 did not require the ABA signaling, although their Psm ES4326-induced expression depends on ABA signaling pathway (Zheng et al. 2012).
Regulation of NAC TF activity
miRNA-mediated regulation of NAC transcript abundance
miRNAs are small, single-stranded, noncoding RNAs that can fine-tune the transcript abundance of their target genes and thus play roles in plant immunity (Seo et al. 2013). Bioinformatics and experimental analyses revealed some NAC genes are targets of several conserved miRNAs and miRNA-mediated regulation of NAC gene transcript abundance is one of the mechanisms that govern the activity of NAC TFs in plant cells. Arabidopsis NAC4 (ANAC079/080), one of the six targets of miR164, was found to promote pathogen-induced cell death under negative regulation by miR164 (Lee et al. 2017). The NAC4-overexpressing plants and the mir164a, mir164b and mir164c mutant plants showed noticeably enhanced cell death after inoculation with an avirulent strain of Pst DC3000 (AvrRpm1). The expression kinetics of NAC4 and miR164 after infection with Pst DC3000 (AvrRpm1), and NAC4-miR164 cleavage assays in protoplasts, demonstrated that NAC4 expression is fine-tuned by the negative action of miR164 through specific cleavage of the NAC4 mRNA during NAC4 promotion of HR cell death (Lee et al. 2017). In rice, miR164a suppresses the expression of OsNAC60, a blast fungus-inducible NAC TF, and overexpression of miR164a leads to enhanced susceptibility to M. oryzae (Wang et al. 2018a). It was also observed that TaNAC21/22 and tae-miR164 showed contrasting divergent expression patterns in wheat in response to Puccinia striiformis f. sp. tritici and that tae-miR164 cleaved the mRNA of TaNAC21/22 genes when both TaNAC21/22 and tae-miR164 genes were transiently co-expressed in tobacco leaves (Feng et al. 2014).
Post-translational modification of NAC proteins
Post-translational modifications such as phosphorylation and ubiquitination can affect the activity, distribution and abundance of proteins in plant cells and have been implicated in plant immunity (Meng and Zhang 2013; Zhou and Zeng 2017). In rice, nuclear localization of the PCD-related OsNAC4 was found to be critical for PCD. OsNAC4 was translocated into nuclei after infection by an avirulent strain of Acidovorax avenae; however, the amount of OsNAC4 accumulation in nuclei was decreased with the absence of staurosporine, a potent inhibitor of serine/threonine protein kinases, indicating that the translocation of OsNAC4 into the nucleus might be regulated by phosphorylation (Kaneda et al. 2009). The tomato SlNAC1, which interacts with the geminivirus replication enhance (REn) protein of Tomato leaf curl virus, was required for tomato immunity against bacterial pathogens (Selth et al. 2005; Huang et al. 2013), and was found to be stable in the presence of proteasome-specific inhibitor MG132 or MG115, indicating ubiquitination of SlNAC1 in plant cells (Huang et al. 2013). Recently, it was found that NAC1 specifically interacts with SINA3, a member of SEVEN IN ABSENTIA family, in the nucleus of plant cells and that SINA3 is a functional ubiquitin E3 ligase capable of promoting NAC1 degradation in planta via polyubiquitination (Miao et al. 2016). It is likely that the function of SlNAC1 in tomato immunity is fine-tuned by SINA5. In addition, the rice PCD-related RIM1 was destabilized by JA treatment and degraded by a 26S proteasome complex (Yoshii et al. 2010).
Interactions with other proteins
Many TFs exert their biochemical functions through interactions with other proteins including other types of TFs to coordinately regulate the expression of their target genes or enhance their binding capacity to the cis-elements in promoters of their target genes (Puranik et al. 2012). The Arabidopsis CBNAC/NTL9, a transcriptional repressor (Kim et al. 2007b), physically interacted with SNI1, a negative regulator of PR1 expression. SNI1 enhanced the DNA-binding activity of the CBNAC/NTL9 to the cis-elements in promoter of PR1, indicating that CBNAC/NTL9 acts synergistically with SNI1 as a transcriptional repressor of PR1 and thus functions as a negative regulator of Arabidopsis immunity (Kim et al. 2012). Another, CBNAC/NTL9 interacts with CRWN1, a member of CROWDED NUCLEI family that are considered as lamin-like candidates. The interaction between CRWN1 and CBNAC/NTL9 enhanced the binding of CBNAC/NTL9 to the promoter of the PR1 gene, increased the repressive function of CBNAC/NTL9 on PR1 expression (Guo et al. 2017). Furthermore, the ANAC019/ANAC055/ANAC072 suppressed whereas TCP8 and TCP9, two members of the TEOSINTE BRANCHED1, CYCLOIDEA and PCF (TCP) family TFs, enhanced ICS1 expression during pathogen infection (Zheng et al. 2012; Wang et al. 2015b). ANAC019 interacted with TCP8 and this interaction may be involved in the orchestrated regulation of ICS1 expression to regulate SA accumulation (Wang et al. 2015b).
Target genes regulated by immunity-related NACs
Transcriptional activity
Most of the NAC TFs functionally characterized have been shown to possess transcriptional activator function. Arabidopsis ATAF1, NTL6, rice OsNAC6, ONAC122, ONAC131, OsNAC111 and RIM1, barley HvNAC6, tomato SlSRN1, and cotton GhATAF1 are transcriptional activators in yeast or in plant protoplasts (Jensen et al. 2007; Lu et al. 2007; Nakashima et al. 2007; Yoshii et al. 2009; Seo et al. 2010; Sun et al. 2013; Yokotani et al. 2014; He et al. 2016). Interestingly, wheat TaNAC1 does not display transcriptional activity although it has a transcription activation domain in its C-terminal (Wang et al. 2015a). On the other hand, a few of the NAC proteins with known biological functions are transcriptional repressors. The Arabidopsis CBNAC/NTL9 and NAC4 function as transcriptional repressors (Kim et al. 2007b; Lee et al. 2017). Notably, previous studies showed that ANAC019, ANAC055 and ANAC072 had transactivation activity (Tran et al. 2004; Bu et al. 2008); however, a recent study indicated that these three NAC TFs may also function as transcriptional suppressors by binding to the ICS1 promoter and repressing its expression (Zheng et al. 2012). This dual transcriptional capability of NAC TFs, functioning either as activators or repressors, may be achieved by recruiting or interacting with different transcriptional partners.
DNA binding sites
As TFs, NAC proteins exert their biological functions through binding to their target sites in promoters of downstream target genes and thus drive the transcription of the target genes. A NAC recognition sequence (NACRS), containing CATGT and harboring CACG, was characterized as a conserved core binding site for some stress-responsive NACs across plant species (Tran et al. 2004) and single NACRS could be enough to drive the binding of NAC TFs to promoters of target genes (Lindemose et al. 2014). The immunity-related ATAF1, ANAC019, ANAC055 and ANAC072 have been shown to be capable of binding to this NACRS core site in stress-responsive genes (Tran et al. 2004; Lindemose et al. 2014). In vitro binding assays revealed that ANAC019 bound directly to the CATGT and CACG motifs in the promoter of the defense gene VSP1 (Bu et al. 2008). The flanking bases next to the core CGT[G/A] of NACRS in promoters may determine the binding specificities and fine-tune affinity for different NAC TFs in vivo (Lindemose et al. 2014). The Arabidopsis CBNAC/NTL9 is capable of binding to a calmodulin-binding NAC binding sequence (CBNACBS) that consists of a GCTT core sequence (Kim et al. 2007b), whereas Arabidopsis NAC4 contains a 9 bp consensus binding sequence ACAAGCAAC (Lee et al. 2017). Genomic pull-down assays identified a 25 bp specific A/T-rich consensus binding sequence with repeating [GC]AAA motifs for ATAF2 and transcriptional analyses confirmed this binding sequence is enough to promote ATAF2-mediated transcription of genes whose promoters contain this consensus sequence (Wang and Culver 2012). According to the current knowledge, it is likely that immunity-related NAC TFs have diverse DNA binding sites to regulate the transcription of the downstream target genes and therefore exert their biological functions in plant immunity.
Target genes
Experimental identification of DNA binding sequences, bioinformatics analyses of the presence of DNA binding sites in gene promoters and comparative analyses of co-expression patterns, along with ChIP verification of in vivo DNA binding events, enable the characterization of putative target genes whose transcription is regulated by specific immunity-related NAC TFs (Table 1). In Arabidopsis, SA biosynthesis genes ICS1, PAD4, EDS1 and PBS3 are the targets for ANAC019/ANAC055/ANAC072 (Zheng et al. 2012) while ICS1 and defense gene PR1 are targets of CBNAC/NTL9 (Kim et al. 2012; Zheng et al. 2015). Microarray, ChIP and qRT-PCR analyses identified at least three NAC4-targeted genes LURP1, WRKY40 and WRKY54, which act as negative regulators of cell death (Lee et al. 2017). The active form of Arabidopsis NTL6 bound to a region of − 105 to − 76 bp in PR1 promoter in vitro and induced the expression of PR1, PR2 and PR5 by directly binding to a conserved sequence in their promoters (Seo et al. 2010) while the rice OsNAC111 was shown to be capable of enhancing the promoter activity of defense genes PR2 and PR8 (Yokotani et al. 2014). Microarray analysis revealed that many abiotic and biotic stress inducible genes were upregulated in OsNAC6-overexpressing rice plants. Among these upregulated stress inducible genes, LOC_Os01g73200 (AK104277, coding for a cationic peroxidase) and LOC_Os04g25060 (AK110725, coding a protein containing a conserved DUF26 domain) were identified as putative target genes of OsNAC6 through transient transactivation assays (Nakashima et al. 2007). In tomato, the JA2L targets the SA metabolism genes SAMT1 and SAMT2 by binding to the NAC core-binding sites in their promoters; whereas JA2 specifically interacted with the NAC core binding sites of the NCED1 promoter and selectively activated the expression of NCED1 (Du et al. 2014). However, the target genes of other reported immunity-related NAC TFs remain to be characterized.
Conclusions and further directions
During the last two decades, functional studies have established our current knowledge on the functions of dozens of NAC TFs as critical regulators of various aspects of plant immunity through impacts on different immune signaling and as virulence targets of pathogen effectors to suppress host plant immunity or benefit pathogen replication. Functions of these immunity-related NAC TFs and their mechanisms of action provide new insights into our understanding of the regulation of plant immunity against pathogens. Further studies should focus on the following three aspects. First, the discovery of novel immunity-related NAC TFs. As a relatively large family of TFs, family-wide functional studies using knockout/knockdown and overexpression approaches will identify novel immunity-related NAC TFs, with emphasis on the NAC TFs in economically important crop plants. For this aspect, CRES-T, tag line, RNAi, and antisense technologies are useful to analyze the biological functions of NAC TFs. For example, the chimeric repressor silencing technology CRES-T system, which is achieved by fusing the NAC proteins with the SRDX repression domain (for SUPERMAN repression domain X), successfully identified some immunity-related NAC TFs such as ATAF1, OsNAC111 and JA2. Second, studies are needed for in-depth understanding of the regulatory network of immunity-related NAC TFs. Further biochemical, genetic and molecular studies will be helpful to elucidate the detailed mechanisms for the immunity-related NAC TFs in transcriptional regulation of their target genes and how these immunity-related NAC TFs integrate into the well-established immune signaling network. Another, global mapping of the genome-wide DNA-binding sites and specificities by high-throughput approaches will discover and characterize the regulatory networks for the immunity-related NAC TFs. Third, emphasis should be given to the potential application of immunity-related NAC TFs. The NAC TFs possess a great potential in improvement of disease resistance in economically important crops. This can be achieved by directly manipulating the expression of the immunity-related NAC TFs via overexpression or knockout/knockdown approaches and by modifying the binding sites in promoters of the immunity-related NAC TFs-regulated target genes through CRISPR/Cas9 genome editing. Notably, some immunity-related NAC TFs such as ATAF1, ATAF2 and CBNAC/NTL9 show distinct and even opposite functions in immunity against necrotrophic and biotrophic pathogens in dicot and monocot plants. Thus, risks from pleiotropic effect of NAC TFs on plant immunity against different pathogens as well as on abiotic stress tolerance and agronomic traits must be taken into consideration when immunity-related NAC TFs are used for genetic improvement of disease resistance in crop plants.
Abbreviations
- AP2/ERF:
-
Apetala2/ethylene responsive factor
- Bgh :
-
Blumeria graminis f. sp. hordei
- Bgt :
-
Blumeria graminis f. sp. tritici
- bHLH:
-
Basic helix-loop-helix
- bZIP:
-
Basic leucine zipper domain
- CAMTA:
-
CaM-binding transcription activator
- CBNAC:
-
Calmodulin-binding NAC protein
- CBNACBS:
-
Calmodulin-binding NAC binding sequence
- CF:
-
Culture filtrate
- CMV:
-
Cucumber mosaic virus
- ER:
-
Endoplasmic reticulum
- ETI:
-
Effector-triggered immunity
- GRAB:
-
Geminivirus Rep A-binding
- hpi:
-
Hours post inoculation
- HR:
-
Hypersensitive response
- JUB1:
-
JUNGBRUNNEN1
- MYMIV:
-
Mungbean yellow mosaic India virus
- NAC:
-
NAM, ATAF and CUC
- NACRS:
-
NAC recognition sequence
- NF-Y:
-
Nuclear Factor Y
- NTL:
-
NAC with transmembrane motif1-like
- OE:
-
Overexpression
- PCD:
-
Programmed cell death
- PTI:
-
PAMP/MAMP/DAMP-triggered immunity
- RDV:
-
Rice dwarf virus
- RNAi:
-
RNA interference
- TCV:
-
Turnip crinkle virus
- TF:
-
Transcription factor
- TIP:
-
TCV-interacting protein
- TLCV:
-
Tomato leaf curl virus
- TM:
-
Transmembrane
- TMV:
-
Tobacco mosaic virus
- VIGS:
-
Virus-induced gene silencing
- WDV:
-
Wheat dwarf geminivirus
References
Arnaud D, Hwang I. A sophisticated network of signaling pathways regulates stomatal defenses to bacterial pathogens. Mol Plant. 2015;8:566–81.
Bigeard J, Colcombet J, Hirt H. Signaling mechanisms in pattern-triggered immunity (PTI). Mol Plant. 2015;8:521–39.
Birkenbihl RP, Liu S, Somssich IE. Transcriptional events defining plant immune responses. Curr Opin Plant Biol. 2017;38:1–9.
Block A, Toruno TY, Elowsky CG, Zhang C, Steinbrenner J, Beynon J, et al. The Pseudomonas syringae type III effector HopD1 suppresses effector-triggered immunity, localizes to the endoplasmic reticulum, and targets the Arabidopsis transcription factor NTL9. New Phytol. 2014;201:1358–70.
Boller T, He SY. Innate immunity in plants: An arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science. 2009;324:742–4.
Bollhoner B, Prestele J, Tuominen H. Xylem cell death: emerging understanding of regulation and function. J Exp Bot. 2012;63:1081–94.
Bu Q, Jiang H, Li CB, Zhai Q, Zhang J, Wu X, et al. Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Res. 2008;18:756–67.
Buscaill P, Rivas S. Transcriptional control of plant defence responses. Curr Opin Plant Biol. 2014;20:35–46.
Chen YJ, Perera V, Christiansen MW, Holme IB, Gregersen PL, Grant MR, et al. The barley HvNAC6 transcription factor affects ABA accumulation and promotes basal resistance against powdery mildew. Plant Mol Biol. 2013;83:577–90.
Coll NS, Epple P, Dangl JL. Programmed cell death in the plant immune system. Cell Death Differ. 2011;18:1247–56.
Cui H, Tsuda K, Parker JE. Effector-triggered immunity: from pathogen perception to robust defense. Annu Rev Plant Biol. 2015;66:487–511.
Delessert C, Kazan K, Wilson IW, Van Der Straeten D, Manners J. Dennis ES, et al the transcription factor ATAF2 represses the expression of pathogenesis-related genes in Arabidopsis. Plant J. 2005;43:745–57.
Donze T, Qu F, Twigg P, Morris TJ. Turnip crinkle virus coat protein inhibits the basal immune response to virus invasion in Arabidopsis by binding to the NAC transcription factor TIP. Virology. 2014;449:207–14.
Du M, Zhai Q, Deng L, Li S, Li H, Yan L, et al. Closely related NAC transcription factors of tomato differentially regulate stomatal closure and reopening during pathogen attack. Plant Cell. 2014;26:3167–84.
Ernst HA, Olsen AN, Larsen S, Lo LL. Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 2004;5:297–303.
Fang Y, You J, Xie K, Xie W, Xiong L. Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol Gen Genomics. 2008;280:547–63.
Feng H, Duan X, Zhang Q, Li X, Wang B, Huang L, et al. The target gene of tae-miR164, a novel NAC transcription factor from the NAM subfamily, negatively regulates resistance of wheat to stripe rust. Mol Plant Pathol. 2014;15:284–96.
Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol. 2005;43:205–27.
Guo T, Mao X, Zhang H, Zhang Y, Fu M, Sun Z, et al. Lamin-LIKE proteins negatively regulate plant immunity through NAC WITH TRANSMEMBRANE MOTIF1-LIKE9 and NONEXPRESSOR OF PR GENES1 in Arabidopsis thaliana. Mol Plant. 2017;10:1334–48.
He X, Zhu L, Xu L, Guo W, Zhang X. GhATAF1, a NAC transcription factor, confers abiotic and biotic stress responses by regulating phytohormonal signaling networks. Plant Cell Rep. 2016;35:2167–79.
Huang PY, Catinot J, Zimmerli L. Ethylene response factors in Arabidopsis immunity. J Exp Bot. 2016;67:1231–41.
Huang W, Miao M, Kud J, Niu X, Ouyang B, Zhang J, et al. SlNAC1, a stress-related transcription factor, is fine-tuned on both the transcriptional and the post-translational level. New Phytol. 2013;197:1214–24.
Hussey SG, Mizrachi E, Creux NM, Myburg AA. Navigating the transcriptional roadmap regulating plant secondary cell wall deposition. Front Plant Sci. 2013;4:325.
Jensen MK, Hagedorn PH, de Torres-Zabala M, Grant MR, Rung JH, Collinge DB, et al. Transcriptional regulation by a NAC (NAM-ATAF1,2-CUC2) transcription factor attenuates ABA signalling for efficient basal defence towards Blumeria graminis f. sp. hordei in Arabidopsis. Plant J. 2008;56:867–80.
Jensen MK, Kjaersgaard T, Nielsen MM, Galberg P, Petersen K, O'Shea C, et al. The Arabidopsis thaliana NAC transcription factor family: structure-function relationships and determinants of ANAC019 stress signalling. Biochem J. 2010;426:183–96.
Jensen MK, Rung JH, Gregersen PL, Gjetting T, Fuglsang AT, Hansen M, et al. The HvNAC6 transcription factor: a positive regulator of penetration resistance in barley and Arabidopsis. Plant Mol Biol. 2007;65:137–50.
Jeong RD, Chandra-Shekara AC, Kachroo A, Klessig DF, Kachroo P. HRT-mediated hypersensitive response and resistance to Turnip crinkle virus in Arabidopsis does not require the function of TIP, the presumed guardee protein. Mol Plant-Microbe Interact. 2008;21:1316–24.
Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–9.
Kaneda T, Taga Y, Takai R, Iwano M, Matsui H, Takayama S, et al. The transcription factor OsNAC4 is a key positive regulator of plant hypersensitive cell death. EMBO J. 2009;28:926–36.
Kazan K, Manners JM. MYC2: the master in action. Mol Plant. 2013;6:686–703.
Kim HJ, Nam HG, Lim PO. Regulatory network of NAC transcription factors in leaf senescence. Curr Opin Plant Biol. 2016;33:48–56.
Kim HS, Park BO, Yoo JH, Jung MS, Lee SM, Han HJ, et al. Identification of a calmodulin-binding NAC protein as a transcriptional repressor in Arabidopsis. J Biol Chem. 2007b;282:36292–302.
Kim HS, Park HC, Kim KE, Jung MS, Han HJ, Kim SH, et al. A NAC transcription factor and SNI1 cooperatively suppress basal pathogen resistance in Arabidopsis thaliana. Nucleic Acids Res. 2012;40:9182–92.
Kim SY, Kim SG, Kim YS, Seo PJ, Bae M, Yoon HK, et al. Exploring membrane-associated NAC transcription factors in Arabidopsis: implications for membrane biology in genome regulation. Nucleic Acids Res. 2007a;35:203–13.
Lai Z, Mengiste T. Genetic and cellular mechanisms regulating plant responses to necrotrophic pathogens. Curr Opin Plant Biol. 2013;16:505–12.
Le Hénanff G, Profizi C, Courteaux B, Rabenoelina F, Gérard C, Clément C, et al. Grapevine NAC1 transcription factor as a convergent node in developmental processes, abiotic stresses, and necrotrophic/biotrophic pathogen tolerance. J Exp Bot. 2013;64:4877–93.
Lee MH, Jeon HS, Kim HG, Park OK. An Arabidopsis NAC transcription factor NAC4 promotes pathogen-induced cell death under negative regulation by microRNA164. New Phytol. 2017;214:343–60.
Li B, Meng X, Shan L, He P. Transcriptional regulation of pattern-triggered immunity in plants. Cell Host Microbe. 2016;19:641–50.
Liang M, Li H, Zhou F, Li H, Liu J, Hao Y, et al. Subcellular distribution of NTL transcription factors in Arabidopsis thaliana. Traffic. 2015;16:1062–74.
Lievens L, Pollier J, Goossens A, Beyaert R, Staal J. Abscisic acid as pathogen effector and immune regulator. Front Plant Sci. 2017;8:587.
Lindemose S, Jensen MK, Van de Velde J, O'Shea C, Heyndrickx KS, Workman CT, et al. A DNA-binding-site landscape and regulatory network analysis for NAC transcription factors in Arabidopsis thaliana. Nucleic Acids Res. 2014;42:7681–93.
Liu B, Ouyang Z, Zhang Y, Li X, Hong Y, Huang L, et al. Tomato NAC transcription factor SlSRN1 positively regulates defense response against biotic stress but negatively regulates abiotic stress response. PLoS One. 2014;9:e102067.
Liu Q, Yan S, Huang W, Yang J, Dong J, Zhang S, et al. NAC transcription factor ONAC066 positively regulates disease resistance by suppressing the ABA signaling pathway in rice. Plant Mol Biol. 2018;98:289–302.
Lu PL, Chen NZ, An R, Su Z, Qi BS, Ren F, et al. A novel drought-inducible gene, ATAF1, encodes a NAC family protein that negatively regulates the expression of stress-responsive genes in Arabidopsis. Plant Mol Biol. 2007;63:289–305.
Lv Z, Wang S, Zhang F, Chen L, Hao X, Pan Q, et al. Overexpression of a novel NAC domain-containing transcription factor (AaNAC1) enhances the content of artemisinin and increases tolerance to drought and Botrytis cinerea in Artemisia annua. Plant Cell Physiol. 2016;57:1961–71.
McLellan H, Boevink PC, Armstrong MR, Pritchard L, Gomez S, Morales J, et al. An RxLR effector from Phytophthora infestans prevents re-localisation of two plant NAC transcription factors from the endoplasmic reticulum to the nucleus. PLoS Pathog. 2013;9:e1003670.
Meng X, Zhang S. MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol. 2013;51:245–66.
Miao M, Niu X, Kud J, Du X, Avila J, Devarenne TP, et al. The ubiquitin ligase SEVEN IN ABSENTIA (SINA) ubiquitinates a defense-related NAC transcription factor and is involved in defense signaling. New Phytol. 2016;211:138–48.
Mukhtar MS, Carvunis AR, Dreze M, Epple P, Steinbrenner J, Moore J, et al. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science. 2011;333:596–601.
Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. NAC transcription factors in plant abiotic stress responses. Biochim Biophys Acta. 2012;1819:97–103.
Nakashima K, Tran LS, Van Nguyen D, Fujita M, Maruyama K, Todaka D, et al. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007;51:617–30.
Noman A, Liu Z, Aqeel M, Zainab M, Khan MI, Hussain A, et al. Basic leucine zipper domain transcription factors: the vanguards in plant immunity. Biotechnol Lett. 2017;39:1779–91.
Nuruzzaman M, Manimekalai R, Sharoni AM, Satoh K, Kondoh H, Ooka H, et al. Genome-wide analysis of NAC transcription factor family in rice. Gene. 2010;465:30–44.
Nuruzzaman M, Sharoni AM, Kikuchi S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front Microbiol. 2013;4:248.
Olsen AN, Ernst HA, Leggio LL, Skriver K. NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 2005;10:79–87.
Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003;10:239–47.
Park SR, Kim HS, Lee KS, Hwang DJ, Bae SC, Ahn IP, et al. Overexpression of rice NAC transcription factor OsNAC58 on increased resistance to bacterial leaf blight. J Plant Biotechnol. 2017;44:149–55.
Peng Y, van Wersch R, Zhang Y. Convergent and divergent signaling in PAMP-triggered immunity and effector-triggered immunity. Mol Plant-Microbe Interact. 2018;31:403–9.
Phukan UJ, Jeena GS, Shukla RK. WRKY transcription factors: molecular regulation and stress responses in plants. Front Plant Sci. 2016;7:760.
Pieterse CM, Leon-Reyes A, Van der Ent S, Van Wees SC. Networking by small-molecule hormones in plant immunity. Nat Chem Biol. 2009;5:308–16.
Pieterse CM, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SC. Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol. 2012;28:489–521.
Puranik S, Sahu PP, Srivastava PS, Prasad M. NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 2012;17:369–81.
Ren T, Qu F, Morris TJ. The nuclear localization of the Arabidopsis transcription factor TIP is blocked by its interaction with the coat protein of Turnip crinkle virus. Virology. 2005;331:316–24.
Ren T, Qu F, Morris TJ. HRT gene function requires interaction between a NAC protein and viral capsid protein to confer resistance to turnip crinkle virus. Plant Cell. 2012;12:1917–25.
Ricachenevsky FK, Menguer PK, Sperotto RA. kNACking on heaven's door: how important are NAC transcription factors for leaf senescence and Fe/Zn remobilization to seeds? Front Plant Sci. 2013;4:226.
Saga H, Ogawa T, Kai K, Suzuki H, Ogata Y, Sakurai N, et al. Identification and characterization of ANAC042, a transcription factor family gene involved in the regulation of camalexin biosynthesis in Arabidopsis. Mol Plant-Microbe Interact. 2012;25:684–96.
Schwessinger B, Ronald PC. Plant innate immunity: perception of conserved microbial signatures. Annu Rev Plant Biol. 2012;63:451–82.
Segonzac C, Zipfel C. Activation of plant pattern-recognition receptors by bacteria. Curr Opin Microbiol. 2011;14:54–61.
Selth LA, Dogra SC, Rasheed MS, Healy H, Randles JW, Rezaian MA. A NAC domain protein interacts with tomato leaf curl virus replication accessory protein and enhances viral replication. Plant Cell. 2005;17:311–25.
Seo JK, Wu JG, Lii YF, Li Y, Jin HL. Contribution of small RNA pathway components in plant immunity. Mol Plant-Microbe Interact. 2013;26:617–25.
Seo PJ, Kim MJ, Park JY, Kim SY, Jeon J, Lee YH, et al. Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J. 2010;61:661–71.
Shahnejat-Bushehri S, Nobmann B, Devi Allu A, Balazadeh S. JUB1 suppresses Pseudomonas syringae-induced defense responses through accumulation of DELLA proteins. Plant Signal Behav. 2016a;11:e1181245.
Shahnejat-Bushehri S, Tarkowska D, Sakuraba Y, Balazadeh S. Arabidopsis NAC transcription factor JUB1 regulates GA/BR metabolism and signalling. Nat Plants. 2016b;2:16013.
Shao HB, Wang HY, Tang XL. NAC transcription factors in plant multiple abiotic stress responses: progress and prospects. Front Plant Sci. 2015;6:902.
Shigenaga AM, Argueso CT. No hormone to rule them all: interactions of plant hormones during the responses of plants to pathogens. Semin Cell Dev Biol. 2016;56:174–89.
Spoel SH, Dong XN. How do plants achieve immunity? Defence without specialized immune cells. Nat Rev Immunol. 2012;12:89–100.
Sun L, Zhang H, Li D, Huang L, Hong Y, Ding XS, et al. Functions of rice NAC transcriptional factors, ONAC122 and ONAC131, in defense responses against Magnaporthe grisea. Plant Mol Biol. 2013;81:41–56.
Suyal G, Rana VS, Mukherjee SK, Wajid S, Choudhury NR. Arabidopsis thaliana NAC083 protein interacts with Mungbean yellow mosaic India virus (MYMIV) rep protein. Virus Genes. 2014;48:486–93.
Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, et al. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. 2004;16:2481–98.
Tsuda Km Somssich IE. Transcriptional networks in plant immunity. New Phytol. 2015;206:932–47.
Vlot AC, Dempsey DA, Klessig DF. Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol. 2009;47:177–206.
Wang B, Wei J, Song N, Wang N, Zhao J, Kang Z. A novel wheat NAC transcription factor, TaNAC30, negatively regulates resistance of wheat to stripe rust. J Integr Plant Biol. 2018b;60:432–43.
Wang F, Lin R, Feng J, Chen W, Qiu D, Xu S. TaNAC1 acts as a negative regulator of stripe rust resistance in wheat, enhances susceptibility to Pseudomonas syringae, and promotes lateral root development in transgenic Arabidopsis thaliana. Front Plant Sci. 2015a;6:108.
Wang G, Zhang S, Ma X, Wang Y, Kong F, Meng Q. A stress-associated NAC transcription factor (SlNAC35) from tomato plays a positive role in biotic and abiotic stresses. Physiol Plant. 2016a;158:45–64.
Wang W, Yuan Y, Yang C, Geng S, Sun Q, Long L, et al. Characterisation, expression, and functional analysis of a novel NAC gene associated with resistance to Verticillium wilt and abiotic stress in cotton. G3 (Bethesda). 2016b;6:3951–61.
Wang X, Basnayake BM, Zhang H, Li G, Li W, Virk N, et al. The Arabidopsis ATAF1, a NAC transcription factor, is a negative regulator of defense responses against necrotrophic fungal and bacterial pathogens. Mol Plant-Microbe Interact. 2009a;22:1227–38.
Wang X, Culver JN. DNA binding specificity of ATAF2, a NAC domain transcription factor targeted for degradation by Tobacco mosaic virus. BMC Plant Biol. 2012;12:157.
Wang X, Gao J, Zhu Z, Dong X, Wang X, Ren G, et al. TCP transcription factors are critical for the coordinated regulation of isochorismate synthase 1 expression in Arabidopsis thaliana. Plant J. 2015b;82:151–62.
Wang X, Goregaoker SP, Culver JN. Interaction of the Tobacco mosaic virus replicase protein with a NAC domain transcription factor is associated with the suppression of systemic host defenses. J Virol. 2009b;83:9720–30.
Wang X, Jiang N, Liu J, Liu W, Wang GL. The role of effectors and host immunity in plant-necrotrophic fungal interactions. Virulence. 2014;5:722–32.
Wang Z, Xia Y, Lin S, Wang Y, Guo B, Song X, et al. Osa-miR164a targets OsNAC60 and negatively regulates rice immunity against the blast fungus Magnaporthe oryzae. Plant J. 2018a; 95:584–97.
Wu A, Allu AD, Garapati P, Siddiqui H, Dortay H, Zanor MI, et al. JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell. 2012;24:482–506.
Wu Y, Deng Z, Lai J, Zhang Y, Yang C, Yin B, et al. Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res. 2009;19:1279–90.
Xie Q, Sanz-Burgos AP, Guo H, García JA, Gutiérrez C. GRAB proteins, novel members of the NAC domain family, isolated by their interaction with a geminivirus protein. Plant Mol Biol. 1999;39:647–56.
Yokotani N, Tsuchida-Mayama T, Ichikawa H, Mitsuda N, Ohme-Takagi M, Kaku H, et al. OsNAC111, a blast disease-responsive transcription factor in rice, positively regulates the expression of defense-related genes. Mol Plant-Microbe Interact. 2014;27:1027–34.
Yoon HK, Kim SG, Kim SY, Park CM. Regulation of leaf senescence by NTL9-mediated osmotic stress signaling in Arabidopsis. Mol Cells. 2008;25:438–45.
Yoshii M, Shimizu T, Yamazaki M, Higashi T, Miyao A, Hirochika H, et al. Disruption of a novel gene for a NAC-domain protein in rice confers resistance to Rice dwarf virus. Plant J. 2009;57:615–25.
Yoshii M, Yamazaki M, Rakwal R, Kishi-Kaboshi M, Miyao A, Hirochika H. The NAC transcription factor RIM1 of rice is a new regulator of jasmonate signaling. Plant J. 2010;61:804–15.
Zanetti ME, Rípodas C, Niebel A. Plant NF-Y transcription factors: key players in plant-microbe interactions, root development and adaptation to stress. Biochim Biophys Acta. 2017;1860:645–54.
Zhang J, Zhou JM. Plant immunity triggered by microbial molecular signatures. Mol Plant. 2010;3:783–93.
Zhang L, Zhang F, Melotto M, Yao J, He SY. Jasmonate signaling and manipulation by pathogens and insects. J Exp Bot. 2017;68:1371–85.
Zheng XY, Spivey NW, Zeng W, Liu PP, Fu ZQ, Klessig DF, et al. Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe. 2012;11:587–96.
Zheng XY, Zhou M, Yoo H, Pruneda-Paz JL, Spivey NW, Kay SA, et al. Spatial and temporal regulation of biosynthesis of the plant immune signal salicylic acid. Proc Natl Acad Sci U S A. 2015;112:9166–73.
Zhong R, Lee C, Ye ZH. Evolutionary conservation of the transcriptional network regulating secondary cell wall biosynthesis. Trends Plant Sci. 2010;15:625–32.
Zhou BJ, Zeng LR. Conventional and unconventional ubiquitination in plant immunity. Mol Plant Pathol. 2017;18:1313–30.
Zhou W, Qian C, Li R, Zhou S, Zhang R, Xiao J, et al. TaNAC6s are involved in the basal and broad-spectrum resistance to powdery mildew in wheat. Plant Sci. 2018;277:218–28.
Zhu Z, Shi J, He M, Cao J, Wang Y. Isolation and functional characterization of a transcription factor VpNAC1 from Chinese wild Vitis pseudoreticulata. Biotechnol Lett. 2012;34:1335–42.
Acknowledgements
We thank Dr. Tesfaye Mengiste, Purdue University, for his suggestions and revisions on the manuscript.
Funding
Study in the corresponding author’s lab was supported by the National Key Research and Development Program of China (No. 2016YFD0100600) and by the Fund for Modern Agro-Industry Technology Research System (CARS-26-11).
Availability of data and materials
Not applicable.
Author information
Authors and Affiliations
Contributions
XY and FS wrote the manuscript; HW, JC and DL discussed on some parts of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Yuan, X., Wang, H., Cai, J. et al. NAC transcription factors in plant immunity. Phytopathol Res 1, 3 (2019). https://doi.org/10.1186/s42483-018-0008-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s42483-018-0008-0