Abstract
Background
Fleas of the genus Ctenocephalides serve as vectors for a number of rickettsial zoonoses, including Rickettsia felis. There are currently no published reports of the presence and distribution of R. felis in India, however, the ubiquitous distribution of its vector Ctenocephalides felis, makes it possible that the pathogen is endemic to the region. This study investigates the occurrence of Rickettsia spp. infection in various subspecies of C. felis infesting dogs from urban areas of Mumbai, Delhi and Rajasthan in India.
Methods
Individual fleas collected off 77 stray dogs from Mumbai, Delhi and Rajasthan were screened for Rickettsia spp. by a conventional PCR targeting the ompB gene. Further genetic characterisation of Rickettsia-positive fleas was carried out using nested PCR and phylogenetic analysis of partial DNA sequences of the gltA and ompA genes. Ctenocephalides spp. were morphologically and genetically identified by PCR targeting a fragment of cox1 gene.
Results
Overall, 56/77 fleas (72.7%), including 22/24 (91.7%) from Delhi, 32/44 (72.7%) from Mumbai and 2/9 (22.2%) from Rajasthan were positive for Rickettsia DNA at the ompB gene. Sequences of gltA fragments confirmed the amplification of Rickettsia sp. genotype RF2125. The ompA gene of Rickettsia sp. genotype RF2125 was characterised for the first time and shown 96% identical to R. felis. Three species of Ctenocephalides were identified, with the Ctenocephalides felis orientis being the dominant flea species (69/77; 89.6%) in India, followed by Ctenocephalides felis felis (8/77; 10.4%).
Conclusions
High occurrence of Rickettsia sp. genotype RF2125 in C. felis orientis and the absence of R. felis suggests a specific vector-endosymbiont adaptation and coevolution of the Rickettsia felis-like sp. within subspecies of C. felis.
Similar content being viewed by others
Background
Rickettsioses caused by Rickettsia spp. are zoonotic vector-borne diseases that have a cosmopolitan distribution. In India, infection with epidemic typhus caused by Rickettsia prowazekii [1], scrub typhus caused by Orientia tsutsugamushi [2,3], murine typhus caused by Rickettsia typhi [4], Mediterranean spotted fever caused by Rickettsia conorii [5,6] and infection by Candidatus Rickettsia kellyi [7] have been reported in humans. Clinical signs in humans typically manifest as febrile illness with myalgia, headache, enlarged painful lymph nodes, a cutaneous ‘rash’, eschar (necrosis at the bite site), respiratory, gastrointestinal and/ or neurological abnormalities [7-9].
In recent years, the ubiquitous nature and public health significance of Rickettsia felis, an emerging rickettsial zoonosis that causes flea-borne spotted fever (FSF) has become increasingly apparent. To date, over 100 human cases have been reported worldwide including in Europe, the Americas, the United States of America (USA), Southeast Asia, Africa and the Middle East [10]. The cat flea, Ctenocephalides felis, is the recognised biological vector and infections of R. felis have been reported in over 25 countries spanning five continents, with infection rates ranging from 15% in New Zealand to 81% in New Caledonia [11,12]. More recently, domestic dogs have also been identified as potential natural mammalian reservoirs for R. felis [13,14]. There are currently no published reports of the presence and distribution of R. felis in India, however, its ubiquitous distribution makes it likely that the pathogen is also endemic to the region.
In India, both flea vectors and canine reservoirs live in close proximity to humans in rural and urban communities. India is estimated to have a stray dog population of 25 million [15] and a pet dog population of over 10 million [16]. Visual inspection of stray dogs from urban areas of Delhi, Mumbai and Sikkim reported a prevalence of flea infestation 40.7%, 42.6% and 75.2% respectively [17]. In Rajasthan, 6% of dogs were reported visually infested with fleas (data not shown). Although human infection with R. felis has not been reported in India, it is possible that the non-specific symptoms that mimic other rickettsial or viral infections coupled with the low clinical index of suspicion for FSF, and low availability of specific diagnostic tests such as PCR, culture and R. felis-specific serological tests, make it likely that many human cases are grossly misdiagnosed.
In the present study, we aim to genetically identify and determine the prevalence of Rickettsia spp. in various subspecies of Ctenocephalides spp. collected from stray dogs in urban areas of Delhi, Mumbai and Rajasthan. Morphology and molecular genotyping based on the mtDNA cytochrome c oxidase subunit I (cox1) gene was applied to demonstrate presence of Ctenocephalides felis felis and Ctenocephalides felis orientis and Ctenocephalides canis.
Methods
Flea collection
Fleas were collected by flea combing the coat of 324 stray dogs undergoing sterilisation through animal birth control and rabies vaccination programs in Mumbai (n = 162), Delhi (n = 162) and Rajasthan (n = 150). For further details of methods and prevalence of flea infestation in dogs for Mumbai and Delhi sampling sites, refer to Abd Rani (2011) [18]. All fleas were fixed in 70% ethanol and transported to the University of Queensland and the University of Melbourne for analysis. A total of 77 fleas, each randomly collected from individual stray dogs in the city of Mumbai (n = 44) and Gurgaon in Delhi (n = 24), and all dogs in Jaipur in Rajasthan (n = 9) were selected for identification and Rickettsia spp. screening using PCR. A single C. canis and two C. felis felis voucher specimens fixed in 70% ethanol were sourced from dogs in the Sikkim area, northeast India. Ectoparasite sampling in Delhi and Mumbai was approved by the University of Queensland Animal Ethics Committee. In Rajasthan, ectoparasite sampling was carried out in accordance with the Animal Welfare Act (2011) of India and overseen by Dr Jack Reece, Veterinarian-in-Charge, Help In Suffering, Rajasthan, India.
Flea identification and extraction of DNA
From selected voucher flea species, total DNA was extracted from fleas whilst retaining flea exoskeletons [19,20]. DNA was isolated using Isolate II Genomic DNA kit (BioLine, Australia) as previously described [20]. DNA was eluted into 50 μL of Tris buffer (pH = 8.5) and stored at −20°C. The flea exoskeleton was soaked in 10% KOH for approximately an hour. Exoskeletons were dehydrated using a series of ethanol washes (70%, 80%, 95%, absolute) for 1 hour each, and slide-mounted in Euparal (Ento Supplies, Australia). The slides were donated to the Australian National Insect Collection (ANIC) in Canberra, Australia. Fleas were identified morphologically using a compound microscope with the aid of keys and descriptions [21,22].
Seventy-seven individual fleas were rinsed with PBS for 10 minutes and mechanically crushed using pellet pestles in a 1.5 ml microcentrifuge tube. Genomic DNA was extracted using the DNeasy Blood & Tissue Kit® (Qiagen, Hilden, Germany) according to the manufacturer’s instructions and eluted in 50 μl of AE Buffer. These samples were then subjected to molecular identification using direct sequence comparisons to those deposited on GenBank and screened for Rickettsia spp. using PCR.
Amplification and phylogenetic analysis of the mtDNA cytochrome c oxidase subunit 1 of fleas
A 5′ fragment of the cytochrome c oxidase subunit I (cox1) coding for COX1 protein was PCR amplified using the generic invertebrate amplification primers: LCO1490 (5′-GGT CAA CAA ATC ATA AAG ATA TTG G-3′)/HC02198 (5′-TAA ACT TCA GGG TGA CCA AAA AAT CA-3′) [23] and Cff-F [S0367] (5′-AGA ATT AGG TCA ACC AGG A-3′) and Cff-R [S0368] (5′-GAA GGG TCA AAG AAT GAT GT-3′) [20] or their combination as well as MLepF1 (5′-GCT TTC CCA CGA ATA AAT AAT A-3′) [24] and HC02198 (5′-TAA ACT TCA GGG TGA CCA AAA AAT CA-3′). Reactions of 30 μl contained MyTaq Red Mix (BioLine, Australia), and approximately 1–10 ng of genomic DNA template (~2 μl). Alternatively, 25 μl reactions contained 5× PCR buffer, 200 μmol dNTP, 1.5 mmol MgCl2, 0.5 units of GoTaq polymerase (Promega). Primers were added at a final concentration of 10 pmol. The cycling was as follows (BioLine mix): denaturing at 95°C for 1 min followed by 35 cycles of 95°C for 15 s, 55°C for 15 s, 72°C for 10 s, and a final elongation for 5 min at 72°C. For the alternative PCR, the cycling was as follows (Promega mix): denaturing at 95°C for 2 min followed by 35 cycles of amplification at 95°C for 30 s, 55°C for 30 s and 72°C for 30 s, and a final extension step of 72°C for 5 min. All PCRs were run with a negative control of sterile PCR-grade water. A positive control with flea DNA representing each of species/subspecies morphologically identified and known to amplify at these conditions from a previous study was included in each run [20].
Aliquots of all PCR reactions were subjected to agarose gel electrophoresis to verify product size and the remainder was submitted for sequencing (Macrogen Ltd, Seoul, Korea). The sequences of voucher flea species have been deposited in GenBank (GenBank: KP229378-KP229385).
Individual sequences of the voucher flea specimens were assembled with CLC Main Workbench 6.9 (CLCbio, Denmark). Composition of the nucleotide sequences and phylogenetic analysis were determined using MEGA6.06 [25]. Sequence divergences were calculated using the Kimura 2 parameter distance model.
Amplification of the ompB, gltA and ompA genes of Rickettsia spp.
Individual flea DNA was initially screened for spotted-fever group Rickettsia spp. with previously described conventional PCR targeting a 297-bp region of the rickettsial outer membrane protein B (ompB) gene [13,26]. Randomly selected Rickettsia-positive fleas were further characterised with conventional nested PCRs on more variable loci targeting a 654 bp fragment of gltA and a 879 bp fragment of the ompA genes of R. felis [14,27]. Secondary ompA primers comprising ompA-F2 (5′-CGGTACAATCATTGCAACTGG-3′) and ompA-R2 (5′-GCTATATCTTCAGCAAATAACG-3′) were designed to increase the sensitivity of the PCR by amplification of product from the primary round. PCR conditions of the secondary PCR were identical to that of the primary [27]. To prevent cross-contamination of DNA, DNA extraction, PCR setup, DNA loading for secondary nested PCR and detection of amplicons were carried out in separate laboratories. Negative control using nuclease-free water was included in every PCR run.
Positive PCR products were submitted for DNA sequencing. DNA sequences were analysed using Finch TV 1.4.0 (Geospiza Inc.) and compared with those available in GenBank using the BLAST algorithm (BLAST Basic Local Alignment Search Tool, 2014). DNA sequences were aligned using BioEdit version 7.2.3 [28] with previous published sequences of the gltA and ompA gene of various rickettsiae species sourced from GenBank. Neighbor-joining analyses were conducted with Tamura-Nei parameter distance estimates, and trees constructed using Mega 4.1 software (www.megasoftware.net). Bootstrap analyses were conducted using 1000 replicates. The sequences of both gltA and ompA genes of Rickettsia spp. have been deposited in GenBank (accession no. KP256357-KP256359, KP406620-KP40662, KP687803-KP687805).
Statistical methods
A Fisher’s Exact Test was performed to determine whether an association exists between the proportions of Rickettsia spp. infection among different subspecies of C. felis identified on surveyed dogs using Vassarstats (http://vassarstats.net/tab2x2.html). Odds ratios were calculated to describe the strength of the association.
Results
Overall, 56/77 fleas (72.7%), including 22/24 (91.7%) from Delhi, 32/44 (72.7%) from Mumbai and 2/9 (22.2%) from Rajasthan were positive for Rickettsia spp. at the ompB gene. All negative controls in each PCR run were PCR-negative. Direct alignment of the partial ompB sequences of Rickettsia isolated from all Indian fleas revealed 99.6% similarity to validated R. felis isolate URRWXCal2 (GenBank: CP000053).
Forty-six (12 Delhi, 32 Mumbai and 2 Rajasthan) and 25 (12 Delhi, 11 Mumbai and 2 Rajasthan) fleas that were positive for Rickettsia at the ompB gene were subjected to further PCRs targeting the more variable gltA and ompA genes, respectively. The sequences of the gltA fragments identified in 21 C. felis isolates were 100% identical to each other and to Rickettsia sp. genotype RF2125 (GenBank: AF516333) and 99.8% identical to Candidatus Rickettsia asemboensis (GenBank: JN315968). Neighbour joining analysis based on the alignment of partial gltA sequence provided strong bootstrap support for the placement of five randomly selected representatives of Rickettsia isolated from C. felis into the same cluster as Rickettsia sp. genotype RF2125 (GenBank: AF516333) and Candidatus Rickettsia asemboensis (GenBank: JN315968) (Figure 1).
Sequences of the ompA fragment amplified from 18 C. felis were 100% identical to each other and 96% identical to validated R. felis isolate URRWXCal2 (GenBank: CP000053). Phylogenetic analysis of the ompA gene revealed moderate support for the placement of all isolates of Rickettsia spp. from Indian fleas within a single cluster distinct to validated R. felis isolate URRWXCal2 (GenBank: CP000053) (Figure 2).
Phylogenetic analysis based on the cox1 fragment placed voucher C. felis felis specimens from Sikkim, India within C. felis felis and as a closely related group (two nucleotide polymorphisms across 513 nt) to C. felis felis haplotype 1 from Australia [20]. The sequences of voucher C. felis orientis strains from Delhi clustered within cox1 sequences from C. felis orientis from Thailand (Figure 3) and cox1 sequences from voucher C. felis orientis strains from Mumbai formed a sister group C. felis orientis, that we consider C. felis orientis. All C. felis orientis from Mumbai and Delhi were morphologically consistent with descriptions of C. felis orientis (Figure 4). C. felis orientis formed a sister group with C. canis collected on dogs from Sikkim (Figure 3).
Phylogenetic relationships of Ctenocephalides felis and Ctenocephalides canis based on nucleotide sequence of the mtDNA cox 1. The tree was inferred using the Minimum Evolution method with distances calculated using Kimura 2-parameter method. There were a total of 658 positions in the final dataset. For the tree shown, all ambiguous positions were removed for each sequence pair. The numbers above the branches indicate percentage of 1000 replicate trees in which the associated taxa clustered together in the bootstrap test. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The tree was rooted using Bradiopsylla echidnae mtDNA cox1 sequence (not shown). The scale is in the units of the number of base substitutions per site. Evolutionary analyses were conducted in MEGA6. Flea species is shown on the right and terminal nodes are labelled with their unique identifier and country of origin. Fleas from India are in black boxes followed by the locality where it was collected, a map is shown in the inset.
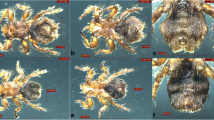
Diagnostic morphological features for differentiation of Ctenocephalides felis felis, Ctenocephalides felis orientis and Ctenocephalides canis . (a) C. felis felis is characterised by a long, acutely angled frons with no part on the vertical plane (white arrow). The dorsal incrassation is long and narrow (black arrow). The posterior margin of the hind tibia of this species has only one notch bearing a stout seta between the longer post-median and apical setae. (b) C. felis orientis is characterised by a short, rounded frons (white arrow) and a shorter dorsal incrassation compared to C. felis felis (black arrow). The posterior margin of the hind tibia is as seen in C. felis felis (black arrow). (c) C. canis is characterised by a short, sharply vertical frons (white arrow) and a short, club-shaped dorsal incrassation. The posterior margin of the hind tibia has two notches bearing stout setae between the post-median and apical setae (black arrows). (d) C. felis orientis can further be distinguished by the presence of a row of tiny setae just dorsal of the antennal fossa in the female (circled), numerous of which are seen in all males of the genus Ctenocephalides.
Ctenocephalides felis species were morphologically identified infesting all dogs in Mumbai, Delhi and Rajasthan (Figures 3, 4). Within C. felis, two subspecies were recognised C. felis orientis and C. felis felis (Figure 4). PCR targeting mtDNA cox1 gene sequence of C. felis using primer pair LCO1490/Cff-R was amplified in 8/77 fleas. The DNA sequences of all eight fleas showed 100% identity to C. felis felis haplotype 3 isolated from Thailand (GenBank: KF684866) and Fiji (GenBank: KF684877) and 98.8% identity to C. felis felis haplotype 1 from Sydney, Australia (KF684882). Fleas that failed to amplify on the first PCR were subjected to a second PCR using primer pair Cff-F/HC02198 that successfully amplified the remaining 69 fleas, of which 22 were subjected to DNA sequencing and identified as C. felis orientis. The dominant flea species infecting dogs in Mumbai, Delhi and Rajasthan was the Oriental cat flea (C. felis orientis) (Table 1).
Statistically, C. felis orientis fleas were 27.5 times more likely to be PCR-positive for ‘Rickettsia sp. genotype RF2125’ than C. felis felis (p = 0.00005).
Discussion
To our knowledge, this is the first study to molecularly characterise rickettsial infection in fleas from India. Previously, C. canis were reported to harbour spotted fever group rickettsiae on the basis of seroconversion in guinea pigs [29]. Dogs are implicated as potential vertebrate reservoir hosts for a number of zoonotic Rickettsia, including R. rickettsii, R. conorii and R. felis [13,30]. The highly ubiquitous nature of fleas and their rickettsial endosymbiont isolated from stray dogs in the current study suggests that this R. felis-like organism (Rickettsia sp. genotype RF2125) may also use dogs as reservoir hosts and pose a potential zoonotic risk to humans. In the current study, a conventional PCR was used to screen the rickettsiae. The distribution of Rickettsia spp. infection may probe higher should a more sensitive molecular assay such as real-time PCR was used in the study.
Since the first detection of R. felis in cat fleas in 1990, this zoonotic rickettsial pathogen has been reported in all continents, except Antarctica. The ubiquitous characteristic of R. felis is associated with the cosmopolitan distribution of the C. felis. Curiously, R. felis was not detected in any of the fleas sourced from stray dogs in this study. The current study provides evidence for the occurrence of Rickettsia sp. genotype RF 2125 as the dominant rickettsiae carried by fleas infesting dogs, with C. felis orientis as the primary carrier. In all cases, the species of Rickettsia detected in fleas in the current study were found to be identical to Rickettsia sp. genotype RF2125, originally detected in a single C. felis (subspecies unknown) and two C. canis isolates near the Thai-Myanmar border [31]. Rickettsia sp. genotype RF2125 was later described in a variety of flea species spanning nine countries -four Archaeopsylla erinacei sourced from hedgehogs in Algeria [32] and two sourced from foxes in France [33]; 12 C. canis isolated from dogs in Gabon [33]; 12 Echidnophaga gallinacea isolated from five black rats in Egypt [34]; a single Pulex irritans sourced from a dog in Hungary [35]; 6/209 C. felis sourced from dogs and cats and from 56/57 rats in Malaysia [36,37]; 2 pools of C. felis from a zookeeper and a grizzly bear in the USA [38], 44/81 C. felis pools sourced from dogs and cats in Costa Rica [39]; and C. felis and C. canis sourced from dogs and cats in Uruguay [40]. The aforementioned studies did not provide detailed morphological or molecular identification of Ctenocephalides spp. to a subspecies level. Ctenocephalides felis is the most common flea in the world with C. felis felis the most widespread subspecies [20]. Other subspecies are more geographically restricted, for example, C. felis damarensis to south western Africa, C. felis strongylus to the Ethiopian zoogeographic region and C. felis orientis to Asia, [20]. C. canis (Curtis) is also widespread but encountered less frequently than C. felis. It has been reported in the USA [41], South America [42], North Africa [43], Europe [44] and Asia [45]. Studies conducted in Thailand [46], north-west Laos and Sabah, Malaysia [47] reported that C. felis orientis was the most common flea species infesting domestic dogs (73.3% - 86.2%), which is comparable to our finding (89.6%; 69/77).
A single study by Kernif et al. (2012) identified species of C. felis sourced from dogs in Laos to a subspecies level [47]. Rickettsial DNA was detected in 69 of 90 (76.6%) fleas. All fleas positive for rickettsial DNA were positive by R. felis-specific qPCR targeting the gltA gene however it is unclear whether conventional PCR and DNA sequencing was carried out on these isolates to confirm their identity as R. felis URRWXCal2. With a mere two base pair difference, it is possible that the qPCR may have cross reacted with Rickettsia sp. genotype RF2125. Interestingly, Kernif et al. (2012) also discovered the frequency of ‘R. felis’ significantly higher in C. felis orientis (59/66; 89.4%) than in C. felis felis (10/19; 52.6%) [47]. The association of the subspecies of cat flea to the species of R. felis-like rickettsiae may be attributed to host-endosymbiont coevolution. A significantly higher prevalence of Rickettsia sp. genotype RF2125 in C. felis orientis compared to C. felis felis in this study suggests that this species of flea could be the primary invertebrate reservoir in India and possibly other parts of Asia where C. felis orientis and R. felis sp. genotype RF2125 co-exist. In addition, in Africa [48] and Europe [49], between 95-100% of hedgehog fleas A. erinacei has been demonstrated as carriers of R. felis. The potential for this flea to also harbour Rickettsia sp. genotype 2125 indicates the potential for hedgehog fleas to act as additional vectors for Rickettsia sp. genotype RF2125. Nevertheless, the absence of R. felis in C. felis orientis sourced from Indian dogs and the absence of R. felis sp. genotype RF2125 from C. felis isolated from Australia, where only C. felis felis is known to occur, raises questions with regard to vector-endosymbiont adaptation and coevolution of the Rickettsia felis-like sp. within subspecies of C. felis.
Genetically, C. felis orientis is more closely related to the dog flea C. canis. C. felis orientis forms a sister group to C. canis (Figure 3) that is phylogenetically distinct to C. felis. Rickettsia sp. genotype RF2125 has been reported in the USA, Central and South America, North Africa and Europe, areas in which C. felis orientis is absent. Given that 12/12 C. canis collected from dogs in Gabon [33] were infected with R. felis sp. genotype RF2125, a co-evolutionary relationship between Rickettsia sp. genotype RF2125 and fleas belonging to the C. canis/C. felis orientis complex is likely and should be explored further.
In addition to characterisation at ompB and gltA gene fragments, a partial region of the ompA gene of R. felis sp. genotype RF2125 was characterised for the first time using published primers designed to be specific to R. felis URRWXCal2 [27]. ompA sequence of this amplified Rickettsia spp. was 96% identical to R. felis URRWXCal2, supporting its potential placement as a new species of Rickettsia [50]. Further demonstration of entire length sequences of other genes such as 16S rRNA and gene D is also required to classify Rickettsia sp. genotype RF2125 as new species [51]. Even though Rickettsia sp. genotype RF2125 has been genetically identified on multiple occasions since 2004, the species has never been isolated in cell culture. Nevertheless, a tentative species should be assigned as a matter of priority.
Conclusion
In conclusion, our study provided the first insight of occurrence of Rickettsia sp. genotype RF2125 infection and its close association with C. felis orientis, the predominant ‘cat flea’ infesting dogs in India. Surveys that include detailed morphological and molecular characterisation of fleas together with their R. felis-like rickettsiae will shed further light on whether host-endosymbiont adaptation is observed in other regions of the world. It is unknown if Rickettsia sp. genotype RF2125 is pathogenic to humans. Nevertheless, this study reveals that the public are at a high risk of exposure to R. felis sp. genotype RF2125 through the bite of C. felis orientis fleas that are ubiquitous on dogs in India.
Abbreviations
- FSF:
-
Flea-borne spotted fever
- qPCR:
-
Real-time polymerase chain reaction
References
Mathai E, Lloyd G, Cherian T, Abraham OC, Cherian AM. Serological evidence for the continued presence of human rickettsioses in southern India. Ann Trop Med Parasitol. 2001;95(4):395–8. doi:10.1080/00034980120065804.
Kumar V, Yadav AK, Iyengar S, Bhalla A, Sharma N, Aggarwal R, et al. Scrub typhus is an under-recognized cause of acute febrile illness with acute kidney injury in India. Plos Neglect Trop D. 2014;8(1):10. doi:10.1371/journal.pntd.0002605.
Dass R, Deka NM, Duwarah SG, Barman H, Hoque R, Mili D, et al. Characteristics of pediatric scrub typhus during an outbreak in the north eastern region of India: peculiarities in clinical presentation, laboratory findings and complications. Indian J Pediatr. 2011;78(11):1365–70. doi:10.1007/s12098-011-0470-5.
Padbidri VS, Kulkarni SM, Joshi MV, Mahadev PVM, Geevarghese G. Murine rickettsiosis along the western ghats of Maharashtra state, India. Indian J Med Res. 1980;71:721–4.
Prakash JAJ, Lal TS, Rosemol V, Verghese VP, Pulimood SA, Reller M, et al. Molecular detection and analysis of spotted fever group Rickettsia in patients with fever and rash at a tertiary care centre in Tamil Nadu. India Pathog Glob Health. 2012;106(1):40–5. doi:10.1179/2047773212y.0000000001.
Tirumala S, Behera B, Jawalkar S, Mishra PK, Patalay PV, Ayyagari S, et al. Indian tick typhus presenting as Purpura fulminans. Indian J Crit Care Med. 2014;18(7):476–8. doi:10.4103/0972-5229.136081.
Rolain JM, Mathai E, Lepidi H, Somashekar HR, Mathew LG, Prakash JAJ, et al. Candidatus Rickettsia kellyi. India Emerg Infect Dis. 2006;12(3):483–5.
Richter J, Fournier PE, Petridou J, Haussinger D, Raoult D. Rickettsia felis infection acquired in Europe and documented by polymerase chain reaction. Emerg Infect Dis. 2002;8(2):207–8.
Rathi N, Rathi A. Rickettsial infections: Indian perspective. Indian Pediatr. 2010;47(2):157–64.
Parola P. Rickettsia felis: from a rare disease in the USA to a common cause of fever in sub-Saharan Africa. Clin Microbiol Infect. 2011;17(7):996–1000. doi:10.1111/j.1469-0691.2011.03516.x.
Kelly PJ, Meads N, Theobald A, Fournier PE, Raoult D. Rickettsia felis, Bartonella henselae, and B. clarridgeiae, New Zealand. Emerg Infect Dis. 2004;10(5):967–8.
Mediannikov O, Cabre O, Qu F, Socolovschi C, Davoust B, Marie JL, et al. Rickettsia felis and Bartonella clarridgeiae in fleas from New Caledonia. Vector Borne Zoonotic Dis. 2011;11(2):181–3. doi:10.1089/vbz.2009.0199.
Hii SF, Kopp SR, Abdad MY, Thompson MF, O’Leary CA, Rees RL, et al. Molecular evidence supports the role of dogs as potential reservoirs for Rickettsia felis. Vector-Borne Zoonot. 2011;11(8):1007–12. doi:10.1089/vbz.2010.0270.
Hii S-F, Kopp SR, Thompson MF, O’Leary CA, Rees RL, Traub RJ. Molecular evidence of Rickettsia felis infection in dogs from northern territory. Australia Parasit Vectors. 2011;4:198. doi:10.1186/1756-3305-4-198.
Sudarshan MK, Mahendra BJ, Madhusudana SN, Ashwoath Narayana DH, Rahman A, Rao NSN, et al. An epidemiological study of animal bites in India: results of a WHO sponsored national multi-centric rabies survey. J Comm Dis. 2006;38(1):32–9.
Pet care in India. Euromonitor International, United Kingdom. 2015. http://www.euromonitor.com/pet-care-in-india/report. Accessed 12 Jan 2015.
Abd Rani PAM. The epidemiology of canine vector-borne diseases in India [PhD thesis]: University of Queensland, School of Veterinary Science. 2012.
Abd Rani PAM, Irwin PJ, Coleman GT, Gatne M, Traub RJ. A survey of canine tick-borne diseases in India. Parasit Vectors. 2011;4:141.
Whiting MF, Whiting AS, Hastriter MW, Dittmar K. A molecular phylogeny of fleas (Insecta: Siphonaptera): origins and host associations. Cladistics. 2008;24(5):677–707. doi:10.1111/j.1096-0031.2008.00211.x.
Lawrence AL, Brown GK, Peters B, Spielman DS, Morin-Adeline V, Slapeta J. High phylogenetic diversity of the cat flea (Ctenocephalides felis) at two mitochondrial DNA markers. Med Vet Entomol. 2014. doi:10.1111/mve.12051.
Dunnet GM, Mardon DK. Monograph of Australian fleas (Siphonaptera). Aust J Zool. 1974;22:1–273.
Hopkins GHE, Eothschild M. An illustrated catalogue of the Rothschild collection of fleas (Siphonaptera) in the British Museum (Natural History): Vol. I. Tungidae and Pulidae. London: Trustees of the British Museum; 1953.
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotech. 1994;3(5):294–9.
Hajibabaei M, Smith MA, Janzen DH, Rodriguez JJ, Whitfield JB, Hebert PDN. A minimalist barcode can identify a specimen whose DNA is degraded. Mol Ecol Notes. 2006;6(4):959–64. doi:10.1111/j.1471-8286.2006.01470.x.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–9. doi:10.1093/molbev/msr121.
Paris DH, Blacksell SD, Stenos J, Graves SR, Unsworth NB, Phetsouvanh R, et al. Real-time multiplex PCR assay for detection and differentiation of rickettsiae and orientiae. Trans R Soc Trop Med Hyg. 2008;102(2):186–93. doi:10.1016/j.trstmh.2007.11.001.
Hii SF, Abdad MY, Kopp SR, Stenos J, Rees RL, Traub RJ. Seroprevalence and risk factors for Rickettsia felis exposure in dogs from Southeast Queensland and the Northern Territory, Australia. Parasit Vectors. 2013;6:159. doi:10.1186/1756-3305-6-159.
Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–8.
Stephen S, Rao KNA. Natural occurrence of spotted-fever group rickettsiae in the dog flea Ctenocephalides canis in Karnataka. Indian J Med Res. 1980;71:870–2.
Levin ML, Killmaster LF, Zemtsova GE. Domestic Dogs (Canis familiaris) as reservoir hosts for Rickettsia conorii. Vector-Borne Zoonot. 2012;12(1):28–33. doi:10.1089/vbz.2011.0684.
Parola P, Sanogo OY, Lerdthusnee K, Zeaiter Z, Chauvancy G, Gonzalez JP, et al. Identification of Rickettsia spp. and Bartonella spp. in fleas from the Thai-Myanmar border. Ann N Y Acad Sci. 2003;990:173–81.
Bitam I, Parola P, De la Cruz KD, Matsumoto K, Baziz B, Rolain JM, et al. First molecular detection of Rickettsia felis in fleas from Algeria. Am J Trop Med Hyg. 2006;74(4):532–5.
Rolain JM, Bitam I, Buffet S, Marie JL, Bourry O, Portelli-Clerc C, et al. Presence or absence of plasmid in Rickettsia felis depending on the source of fleas. Clin Microbiol Infect. 2009;15:296–7. doi:10.1111/j.1469-0691.2008.02245.x.
Loftis AD, Reeves WK, Szumlas DE, Abbassy MM, Helmy IM, Mortarity JR, et al. Surveillance of Egyptian fleas for agents of public health significance: Anaplasma, Bartonella, Coxiella, Ehrlichia, Rickettsia, and Yersinia pestis. Am J Trop Med Hyg. 2006;75(1):41–8.
Hornok S, Meli ML, Perreten A, Farkas R, Willi B, Beugnet F, et al. Molecular investigation of hard ticks (Acari: Ixodidae) and fleas (Siphonaptera: Pulicidae) as potential vectors of rickettsial and mycoplasmal agents. Vet Microbiol. 2010;140(1–2):98–104. doi:10.1016/j.vetmic.2009.07.013.
Mokhtar AS, Tay ST. Short report: molecular detection of Rickettsia felis, Bartonella henselae, and B. clarridgeiae in fleas from domestic dogs and cats in Malaysia. Am J Trop Med Hyg. 2011;85(5):931–3. doi:10.4269/ajtmh. 2011.10-0634.
Tay ST, Mokhtar AS, Low KC, Mohd Zain SN, Jeffery J, Abdul Aziz N, et al. Identification of rickettsiae from wild rats and cat fleas in Malaysia. Med Vet Entomol. 2014;28 Suppl 1:104–8. doi:10.1111/mve.12075.
Nelder MP, Reeves WK, Adler PH, Wozniak A, Wills W. Ectoparasites and associated pathogens of free-roaming and captive animals in zoos of South Carolina. Vector-Borne Zoonot. 2009;9(5):469–77. doi:10.1089/vbz.2008.0008.
Troyo A, Alvarez D, Taylor L, Abdalla G, Calderon-Arguedas O, Zambrano ML, et al. Short report: Rickettsia felis in Ctenocephalides felis from Guatemala and Costa Rica. Am J Trop Med Hyg. 2012;86(6):1054–6. doi:10.4269/ajtmh. 2012.11-0742.
Venzal JM, Perez-Martinez L, Felix ML, Portillo A, Blanco JR, Oteo JA. Prevalence of Rickettsia felis in Ctenocephalides felis and Ctenocephalides canis from Uruguay. Ann N Y Acad Sci. 2006;1078:305–8. doi:10.1196/annals.1374.056.
Durden LA, Judy TN, Martin JE, Spedding LS. Fleas parasitizing domestic dogs in Georgia, USA: species composition and seasonal abundance. Vet Parasitol. 2005;130(1–2):157–62. doi:10.1016/j.vetpar.2005.03.016.
Gonzalez A, Castro DD, Gonzalez S. Ectoparasitic species from Canis familiaris (Linne) in Buenos Aires province. Argentina Vet Parasitol. 2004;120(1–2):123–9. doi:10.1016/j.vetpar.2003.12.001.
Abdel-Mageid MYRAD. Epidemiological study of ectoparasites in stray dogs in Kalubyia Governorate of Egypt with a special reference to its control in puppies by deltamethrin and ivermectin. Lucrari Stiintifice - Medicina Veterinara, Universitatea de Stiinte Agricole si Medicina Veterinara “Ion Ionescu de la Brad” Iasi. 2010;53(12(2)):307–17.
Farkas R, Gyurkovszky M, Solymosi N, Beugnet F. Prevalence of flea infestation in dogs and cats in Hungary combined with a survey of owner awareness. Med Vet Entomol. 2009;23(3):187–94.
Nithikathkul C, Polseela R, Iamsa-ard J, Wongsawad C, Jittapalapong S. A study of ectoparasites of Canis lupus familiaris in Mueang district, Khon Kaen, Thailand. Southeast Asian J Trop Med Public Health. 2005;36 Suppl 4:149–51.
Changbunjong T, Buddhirongawatr R, Suwanpakdee S, Siengsanan J, Yongyuttawichai P, Cheewajorn K, et al. A survey of ectoparasitic arthropods on domestic animals in Tak province, Thailand. Southeast Asian J Trop Med Public Health. 2009;40(3):435–42.
Kernif T, Socolovschi C, Wells K, Lakim MB, Inthalad S, Slesak G, et al. Bartonella and Rickettsia in arthropods from the Lao PDR and from Borneo. Malaysia Comp Immunol Microb. 2012;35(1):51–7. doi:10.1016/j.cimid.2011.10.003.
Khaldi M, Socolovschi C, Benyettou M, Barech G, Biche M, Kernif T, et al. Rickettsiae in arthropods collected from the North African Hedgehog (Atelerix algirus) and the desert hedgehog (Paraechinus aethiopicus) in Algeria. Comp Immunol Microb. 2012;35(2):117–22. doi:10.1016/j.cimid.2011.11.007.
Marie J-L, Davoust B, Socolovschi C, Raoult D, Parola P. Molecular detection of rickettsial agents in ticks and fleas collected from a European hedgehog (Erinaceus europaeus) in Marseilles. France Comp Immunol Microb. 2012;35(1):77–9. doi:10.1016/j.cimid.2011.11.005.
Fournier PE, Roux V, Raoult D. Phylogenetic analysis of spotted fever group rickettsiae by study of the outer surface protein rOmpA. Int J Syst Bacteriol. 1998;48:839–49.
Fournier PE, Dumler JS, Greub G, Zhang JZ, Wu YM, Raoult D. Gene sequence-based criteria for identification of new Rickettsia isolates and description of Rickettsia heilongjiangensis sp nov. J Clin Microbiol. 2003;41(12):5456–65. doi:10.1128/jcm. 41.12.5456-5465.2003.
Acknowledgements
Financial support for this project was provided by the Australian Research Council and Bayer Animal Health. In-kind support through assistance with field-work was provided by the following Non-Government Organisations: Help in Suffering, Jaipur, Jeevaashram and Krishnaashram (Gurgeon, Delhi) and In Defence of Animals (Mumbai). Special thanks to Prof. Gatne, Mumbai Veterinary College, Dr. Vinod Sharma, Jeevasharam and Ms. Aashima Auplish for their assistance in conducting fieldwork. The paper has been sponsored by Bayer Health Care – Animal Health Division in the framework of the 10th CVBD World Forum Symposium. Professor Ian Beveridge is gratefully acknowledged for his assistance with the initial morphological identification of fleas.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SFH carried out laboratory work, data analysis, intellectual interpretation and writing of the manuscript. RJT designed and supervised the study, assisted the field work, provided intellectual interpretation and critical revision of the manuscript for publication. ALL carried out laboratory work, analysis and intellectual interpretation of data. JS was involved in supervision of laboratory work, intellectual interpretation and critical revision of the manuscript for publication. LC and RT assisted in conducting laboratory work. PAMAR carried out the fieldwork and morphological identification of fleas to a species level. All authors read and approved the final version of the manuscript.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Hii, SF., Lawrence, A.L., Cuttell, L. et al. Evidence for a specific host-endosymbiont relationship between ‘Rickettsia sp. genotype RF2125’ and Ctenocephalides felis orientis infesting dogs in India. Parasites Vectors 8, 169 (2015). https://doi.org/10.1186/s13071-015-0781-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-015-0781-x