Abstract
Background
The runt-related transcription factor 1 (RUNX1) gene is a transcription factor that acts as a master regulator of hematopoiesis and represents one of the most frequent targets of chromosomal rearrangements in human leukemias. The t(7;21)(p22;q22) rearrangement generating a 5‘RUNX1-3’USP42 fusion transcript has been reported in two cases of pediatric acute myeloid leukemia (AML) and further in eight adult cases of myeloid neoplasms. We describe the first case of adult AML with a 5‘RUNX1-3’USP42 fusion gene generated by an insertion event instead of chromosomal translocation.
Methods
Conventional and molecular cytogenetic analyses allowed the precise characterization of the chromosomal rearrangement and breakpoints identification. Gene expression analysis was performed by quantitative real-time PCR experiments, whereas bioinformatic studies were carried out for revealing structural genomic characteristics of breakpoint regions.
Results
We identified an adult AML case bearing a ins(21;7)(q22;p15p22) generating a 5‘RUNX1-3’USP42 fusion gene on der(21) chromosome and causing USP42 gene over-expression. Bioinformatic analysis of the genomic regions involved in ins(21;7)/t(7;21) showed the presence of interchromosomal segmental duplications (SDs) next to the USP42 and RUNX1 genes, that may underlie a non-allelic homologous recombination between chromosome 7 and 21 in AML.
Conclusions
We report the first case of a 5‘RUNX1-3’USP42 chimeric gene generated by a chromosomal cryptic insertion in an adult AML patient. Our data revealed that there may be a pivotal role for SDs in this very rare but recurrent chromosomal rearrangement.
Similar content being viewed by others
Background
The runt-related transcription factor 1 (RUNX1) gene is a transcription factor that acts as a master regulator of hematopoiesis and is crucial for the regulation of adult hematopoiesis and differentiation of committed cells of various lineages [1]-[3]. It is one of the most frequent targets of chromosomal rearrangements in human leukemias, in fact, more than 30 chromosomal translocations involving RUNX1 have been described [4]. The t(7;21)(p22;q22) rearrangement was first reported in 2006 in a case of pediatric acute myeloid leukemia (AML) [5]. This kind of chromosomal translocation generates a 5‘RUNX1-3’USP42 fusion transcript, containing the first 5 to 7 exons of RUNX1 fused to exon 3 of ubiquitin specific peptidase 42 (USP42) gene on the der(7) chromosome. To date, the t(7;21)(p22;q22) rearrangement has been reported in another AML child [6], and in nine adult cases of myeloid neoplasms (1 myelodysplastic syndrome and 8 AML) harbouring the 5‘RUNX1-3’USP42 chimeric transcript [7]-[11]. The incidence rate of this rare but recurrent abnormality has been estimated to range from 0.75% to 1% in different adult AML series [9],[10]. Segmental duplications (SDs), accounting for about 10% of the human genome, are DNA sequences larger than 1 kb, found at least twice with a more than 90% sequence similarity in the genome [12]. We report the first case of adult AML with a 5‘RUNX1-3’USP42 fusion gene generated by an insertion event instead of chromosomal translocation. The structural characteristics of the genomic regions involved in ins(21;7)/t(7;21) were revealed by bioinformatic analysis, and the role of SDs in the chromosomal rearrangement is discussed.
Case presentation
A 65-year-old male was admitted to our department with anemia, leukopenia and thrombocytopenia (hemoglobin 8.7 g/dL, total leukocytes 1.48 × 109/L, and platelets 98 × 109/L) and fever. A peripheral blood film showed circulating blast cells (12%). The bone marrow exhibited 44% of myeloid blast cells and immunophenotypic analysis showed that blast cells were positive for HLA-DR, CD4, CD5, CD7, CD13, CD33, CD34, CD11b, CD117 and CD56. Karyotypic analysis revealed 46,XY,t(7;21)(p?15;q22) [12] /46,idem,del(5q)(q22q35) [8] (Figure 1A,B). Molecular analysis did not show NPM1 and FLT3 gene mutations. A diagnosis of AML with maturation (M2 subtype) and “AML NOS” was made according to the FAB and the 2008 WHO criteria, respectively. He was started on an induction treatment regimen containing daunorubicin and cytosine arabinoside but did not obtain complete hematologic remission. After two courses of fludarabine, cytarabine, granulocyte colony-stimulating factor, and idarubicin (FLAG-IDA), complete hematological remission was achieved. At this time banding analysis showed a normal karyotype. The patient started maintainance therapy with azacytidine (75 mg/m2 s.c., day 1 to 7, repeated every 28 days). After 5 cycles of hypomethylating agent the patient is doing well; he is still in hematological and cytogenetic complete remission after eleven months from the diagnosis.
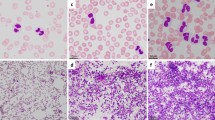
Karyotypic, FISH and molecular analyses in our AML patient with the ins(21;7) rearrangement. (A) GTG-banded karyotype showing the rearrangement between 7 and 21 chromosomes and del(5q) (indicated by arrows) (B) A partial G-banded karyogram comprising both der(7) and der(21) chromosomes (C) FISH analysis with WCP probes specific for chromosomes 7 and 21 showing chromosome 7 insertion on chromosome 21; (D) FISH experiment with the overlapping clones RP11-1006 L1 and RP11-1112A12 and BAC clone RP11-805P12 showed a fusion signal on der(21) identifying ins(21;7) breakpoints; (E) Partial sequence chromatogram showing that RUNX1 exon 7 is fused to USP42 exon 3 in the 5‘RUNX1-3’USP42 transcript. (F) Graphic representation of USP42 gene relative expression using primers specific for wild-type (red), and wild-type and rearranged USP42 gene (blue) in the AML patient with ins(21;7), in the NK-AML pool, and in normal bone marrow (NBM) samples.
Results
Fluorescence in situ hybridization (FISH) analysis with whole chromosome painting (WCP) probes specific for chromosomes 7 and 21 showed the presence of chromosome 7 sequences insertion on chromosome 21, rather than a chromosomal translocation (Figure 1C). Reiterative FISH cohybridizations were performed with 13 and 2 bacterial artificial chromosome (BAC) clones belonging to chromosomes 7 and 21, respectively (Table 1). In details, a chromosome 7 region of about 19 Mb was transferred on chromosome 21; the distal and proximal insertion breakpoints were mapped inside the BAC clone RP11-805P12 and between the clones RP11-813D23 and RP11-592D15, respectively (Table 1; Figure 1D). The chromosome 21 insertion point was localized between the overlapping clones RP11-1006 L1 and RP11-1112A12, generating splitting signals on der(21), due to chromosome 7 insertion (Figure 1D). Therefore, according to FISH data the karyotype was revised as follows: 46,XY,ins(21;7)(q22;p15p22)[12]/46,idem,del(5q)(q22q35)[8].
The UCSC database was queried to identify genes mapping in correspondence with chromosomal breakpoints. The USP42 and cytochrome c (CYCS) genes were mapped next to the distal and proximal chromosome 7 breakpoint regions, respectively; the RUNX1 gene was located at the chromosome 21 insertion point. RT-PCR experiments were carried out to verify the generation of a possible fusion gene. Two chimeric 5‘RUNX1-3’USP42 transcripts were detected, revealing an in-frame fusion of RUNX1 exon 7 with USP42 exon 3 (Figure 1E) and an alternative splice variant missing of RUNX1 exon 6, as reported in previous studies [5]. Molecular analysis performed at the time of complete remission and during the follow up did not reveal the presence of any 5‘RUNX1-3’USP42 fusion transcript. Expression analysis of the USP42 gene was performed as previously reported [10]. Relative expression of the wild-type (using primers spanning exons 2–3) and wild-type and rearranged (using primers spanning exons 5–6) USP42 gene was analyzed in the ins(21;7) AML case and in a pool of samples deriving from four adult normal karyotype AML (NK-AML) and healthy individuals, respectively. USP42 expression was higher in the ins(21;7) sample, whereas the wild type USP42 gene was normally expressed in all specimens (Figure 1F).
Bioinformatic analysis of chromosome 7 and 21 sequences was performed to assess whether the presence of SDs could explain the breakpoints clustering next to the USP42 and RUNX1 genes. Eight paired blocks of interchromosomal SDs (ranging from 3.6 kb to 13.3 kb in size) are localized near the USP42 and RUNX1 genes, at a distance of about 671 Kb and 2.3 Mb, respectively (Figures 2A and 2B); these SDs have a homology ranging from 92% to 95%. In total, the SDs7/21 and SDs21/7 encompass chromosomal regions of 51.9 Kb (chr7:6,872,808-6,963,213) and 43.3 Kb (chr21:33,800,439-33,995,925), respectively. Moreover, it is noteworthy that SDs7/21 are present mostly in the genomic regions adjacent to the USP42 gene breakpoints rather than being homogeneously distributed over the entire chromosome 7. In fact, among the nine SDs7/21 mapped on 7p, 8 were localized near the USP42 gene, whereas among the 16 SDs21/7 identified on 21q, 8 were mapped near the RUNX1 gene (Figure 2A).
Schematic diagram of SDs7/21 and SDs21/7 mediating the chromosomes 7 and 21 rearrangement. (A) SDs7/21 and SDs21/7 distribution along the p and q arms, respectively; SDs located next to USP42 and RUNX1 genes are indicated by square boxes; (B) Detailed genomic organization of SDs7/21 (green) and SDs21/7 (red) adjacent to the USP42 and RUNX1 genes is reported. The ends of the segments that constitute each duplication are indicated by capital letters whereas the horizontal black line represents not duplicated genomic regions. The size of each segment and of single copy sequences is reported in Kb. (C) Hypothetical mechanism at the basis of the 5‘RUNX1-3’USP42 fusion gene generation. The two SDs blocks, SDs7/21 (A-H, in green) and SDs21/7 (A’-H’, in red), promote the approach of chromosomes 7 and 21 and mediate the rearrangement (translocation/insertion) generating the 5‘RUNX1-3’USP42 chimeric gene.
Discussion
We report for the first time a case of adult AML with the 5‘RUNX1-3’USP42 fusion gene generated by a chromosome insertion instead of a translocation mechanism. It is noteworthy that, due to the chromosome 7p bands orientation relative to the der(21) centromere, the generation of this fusion gene can be explained by hypothesizing a direct insertion. The ins(21;7)(q22;p15p22) rearrangement showed three chromosomal breaks: the first two delimited the inserted segment from the donor chromosome 7 and the last one represents the insertion site of the recipient chromosome 21. Moreover, in our case the 5‘RUNX1-3’USP42 fusion gene is localized on der(21) instead of der(7) chromosome as in patients with the t(7;21) translocation. Noteworthy, as previously reported in literature, the reciprocal gene 5‘USP42-3’RUNX1 generated by t(7;21)(p22;q22) results to be an inactive fusion; accordingly, our case showing ins(21;7)(q22;p15p22) without the generation of the reciprocal chimeric gene, underlines that the 5‘USP42- 3’RUNX1 fusion is irrelevant in AML or MDS pathogenesis. Among patients bearing the 5΄RUNX1-3΄USP42 chimeric gene, the mechanism of insertion at the basis of this kind of rearrangement can be estimated to have a frequency of approximately 8%. In this respect, the insertion mechanism rather than translocation represents a rare but probable finding in cases of myeloid neoplasms associated with recurrent and more frequent chimeric genes. For example, PML-RARα, BCR-ABL1 and RUNX1-ETO fusion genes occurred with the insertion mechanism in 2%, 1% and up to 7%, respectively [13]-[15].
The pathogenetic consequences of the 5΄RUNX1-3΄USP42 rearrangement are very hard to define, since the USP42 functions are not known. In fact, only two papers [16],[17] reported a role for the USP42 protein, that is a typical deubiquitinating enzyme, with an important role in mouse embryonic development and in p53 regulation. The 5΄RUNX1-3΄USP42 transcripts encode for a predicted fusion protein retaining the Runt homology domain (RHD), responsible for DNA binding and heterodimerization with core-binding factor β, and the USP42 catalytic ubiquitin carboxyl terminal hydroxylase domain [5]. The leukemogenic effect of the RUNX1-USP42 fusion protein could on one hand be mediated by the dominant-negative inhibitor effect exerted by the RHD domain on the wild-type RUNX1, as reported in other RUNX1 fusion genes [4], and on the other, an impairment of USP42 function could be responsible for the decreased p53 stability.
As already described in the majority of cases with the t(7;21), our ins(21;7) AML case did not achieve complete remission after the first induction treatment, suggesting that the 5΄RUNX1-3΄USP42 rearrangement could identify a category of high-risk AML patients. However, our patient has maintained complete remission after the second line of therapy, thanks to a hypomethylating-based maintenance treatment. Too few data are currently available to assert that leukemic cells bearing the 5΄RUNX1-3΄USP42 rearrangement may be more sensitive to other chemotherapy drugs rather than standard induction treatment.
In terms of SDs distribution across the genome, there are profound differences within chromosomes. Apart from large SD clusters in the subtelomeric and pericentromeric regions of most chromosomes, SDs can also accumulate in interstitial hubs [18]. These hubs are characterized by an increased genomic instability and may be accountable for a non-allelic homologous recombination (NAHR). Therefore, NAHR may be driven by breakpoint-flanking SDs, which can misalign in meiosis due to their sequence homology [19]. Genotype–phenotype relationships for NAHR-mediated rearrangements are well-known, and in recurrent constitutional chromosomal rearrangements are associated with congenital human genomic disorders [19]-[21]. However, the role of SDs in genomic rearrangements associated with cancer is still virtually unknown. In this regard, it has been reported that the isochromosome i(17q) generation in cancer is due to the presence of SDs in correspondence with the chromosomal breakpoints [22]. Moreover, recently our group showed the involvement of SDs in the genesis of the t (9;22) translocation and in the occurrence of genomic deletions on the der(9) chromosome in chronic myeloid leukemia [23]. In our current report we describe the presence of interchromosomal SDs next to USP42 and RUNX1 genes that could be at the basis of NAHR between chromosome 7 and 21 in AML (Figure 2A-C). In fact, the two SDs blocks could promote chromosomes 7 and 21 approach, triggering the NAHR mechanism and the 5΄RUNX1-3΄USP42 fusion gene generation. In this context, it is very hard to identify the circumstances determining the translocation instead of insertion. This observation has already been made [5] but we report for the first time the detailed SDs organization in the genomic region involved in the 5΄RUNX1-3΄USP42 rearrangement (Figure 2A-C). The relationship between the two SDs blocks and the breakpoints on chromosome 7 and 21 appears anything but random considering the fact that the t(7;21)(p22;q22) in AML is a very rare but recurrent rearrangement. A link between nuclear architecture, in terms of chromatin organization, and SDs across the chromosome 7 has been recently demonstrated [24]. What might be the circumstances that favor a chromosome pairing mediated by SDs in AML is something still to be clarified.
Conclusions
We report the first case of a 5΄RUNX1-3΄USP42 chimeric gene generated by a chromosomal cryptic insertion in an adult AML patient. Our data reveal that it is possible that there may be a pivotal role for SDs at the basis of this very rare but recurrent chromosomal rearrangement, including translocation or insertion.
Methods
Cytogenetic analysis
Karyotyping was performed at diagnosis on bone marrow cells according to standard methods. The bone marrow cells were cultured for 24–48 hours, and chromosomes were G-banded with trypsin–Giemsa staining (GTG-banded) according to the recommendations of the International System for Human Cytogenetic Nomenclature [25]. At least 20 metaphases were analyzed.
FISH analysis
FISH analyses were performed on BM samples at the onset of AML, using WCP and BAC probes selected according to the University of California Santa Cruz (UCSC http://genome.ucsc.edu/; Feb. 2009 release) database. Chromosome preparations were hybridized in situ with probes labeled by nick translation [26].
Molecular analyses
Total RNA derived from bone marrow (BM) cells was reverse transcribed into cDNA using the QuantiTect reverse transcription kit (Qiagen, Chatsworth, CA, USA). PCR for detection of the chimaeric 5΄RUNX1-3΄USP42 transcript was performed with the previously reported primers [5]. The amplification was achieved using the following cycling parameters: 95°C for 30 sec, 60°C for 45 sec, and 72°C for 1 min for 35 cycles. Amplification product was run, excised, and extracted from a 2% agarose gel. Products were purified using the QIAquick gel extraction kit (Qiagen) according to the manufacturer’s instructions and sequenced by Sanger sequencing.
Gene expression analysis was carried out by quantitative real-time PCR (qRT-PCR) experiments using the LightCycler 480 SYBR Green I Master mix on the LightCycler 480II (Roche Diagnostics, Indianapolis, IN, USA). All samples were run in triplicate as technical replicates. The β-glucuronidase (β-GUS) gene was employed as endogenous control and a pool of cDNA derived from healthy individuals BM cells was used as calibrator. Amplifications were carried out at 95°C for 10 min, followed by 45 cycles at 95°C (10 sec), 60°C (30 sec), 72°C (1 sec). Advanced relative quantification analysis was performed using LightCycler 480 Software 1.5.1, based on the ∆∆Ct method. For USP42 gene expression analysis the previously reported primers were used [10].
Bioinformatic analysis
The UCSC Table Browser (http://genome.ucsc.edu/cgi-bin/hgTables) was queried using the track `Segmental Dups’ to check for the presence of SDs on chromosomes 7 and 21. The analysis was restricted to the search for interchromosomal SDs with homology for chromosome 21 (SDs7/21, mapped on chromosome 7) and 7 (SDs21/7, mapped on chromosome 21).
Ethics statement
This study was performed in agreement with the Declaration of Helsinki, and approved by the local Ethical Committee (Azienda Ospedaliero Universitaria – Policlinico di Bari) . Written informed consent was obtained from the patient for publication of this Case report and any accompanying images. A copy of the written consent is available for review by the Editor of this journal.
Authors’ contributions
AZ was involved in the execution of the experiments, interpreted data and wrote the manuscript. LA, NC and GT conducted FISH experiments and interpreted data. PC and AC performed conventional cytogenetic analysis; AM, CFM, CB and CC performed molecular and bioinformatics analyses. GS and FA participated in the design of the study and supervised the manuscript preparation. All authors have read and approved the final manuscript.
Abbreviations
- AML:
-
Acute myeloid leukemia
- BAC:
-
Bacterial artificial chromosome
- β-GUS:
-
β-glucuronidase
- BM:
-
Bone marrow
- CYCS :
-
Cytochrome c
- FISH:
-
Fluorescence in situ hybridization
- GTG-banded:
-
G-banded with trypsin–Giemsa staining
- NAHR:
-
Non-allelic homologous recombination
- NBM:
-
Normal bone marrow
- NK-AML:
-
Normal karyotype AML
- qRT-PCR:
-
Quantitative real-time PCR
- RHD:
-
Runt homology domain
- RUNX1:
-
Runt-related transcription factor 1
- SDs:
-
Segmental duplications
- UCSC:
-
University of California Santa Cruz
- USP42:
-
Ubiquitin carboxyl-terminal hydrolase 42
- WCP:
-
Whole chromosome painting
References
Tenen DG, Hromas R, Licht JD, Zhang DE: Transcription factors, normal myeloid development, and leukemia. Blood 1997, 90: 489–519.
Ichikawa M, Goyama S, Asai T, Kawazu M, Nakagawa M, Takeshita M, Chiba S, Ogawa S, Kurokawa M: AML1/Runx1 negatively regulates quiescent hematopoietic stem cells in adult hematopoiesis. J Immunol 2008, 180: 4402–4408. 10.4049/jimmunol.180.7.4402
Link KA, Chou FS, Mulloy JC: Core binding factor at the crossroads: determining the fate of the HSC. J Cell Physiol 2010, 222: 50–56. 10.1002/jcp.21950
De Braekeleer E, Ferec C, De Braekeleer M: RUNX1 translocations in malignant hemopathies. Anticancer Res 2009, 29: 1031–1037.
Paulsson K, Békássy AN, Olofsson T, Mitelman F, Johansson B, Panagopoulos I: A novel and cytogenetically cryptic t(7;21)(p22;q22) in acute myeloid leukemia results in fusion of RUNX1 with the ubiquitin-specific protease gene USP42. Leukemia 2006, 20: 224–229. 10.1038/sj.leu.2404076
Masetti R, Togni M, Astolfi A, Pigazzi M, Indio V, Rivalta B, Manara E, Rutella S, Basso G, Pession A, Locatelli F: Whole transcriptome sequencing of a paediatric case of de novo acute myeloid leukaemia with del(5q) reveals RUNX1-USP42 and PRDM16-SKI fusion transcripts. Br J Haematol 2014, 166: 449–452. 10.1111/bjh.12855
Ji J, Loo E, Pullarkat S, Yang L, Tirado CA: Acute myeloid leukemia with t(7;21)(p22;q22) and 5q deletion: a case report and literature review. Exp Hematol Oncol 2014, 3: 8. 10.1186/2162-3619-3-8
Panagopoulos I, Gorunova L, Brandal P, Garnes M, Tierens A, Heim S: Myeloid leukemia with t(7;21)(p22;q22) and 5q deletion. Oncol Rep 2013, 30: 1549–1552.
Jeandidier E, Gervais C, Radford-Weiss I, Zink E, Gangneux C, Eischen A, Galoisy AC, Helias C, Dano L, Cammarata O, Jung G, Harzallah I, Guérin E, Martzolff L, Drénou B, Lioure B, Tancrédi C, Rimelen V, Mauvieux L: A cytogenetic study of 397 consecutive acute myeloid leukemia cases identified three with a t(7;21) associated with 5q abnormalities and exhibiting similar clinical and biological features, suggesting a new, rare acute myeloid leukemia entity. Cancer Genet 2012, 205: 365–372. 10.1016/j.cancergen.2012.04.007
Giguére A, Hébert J: Microhomologies and topoisomerase II consensus sequences identified near the breakpoint junctions of the recurrent t(7;21)(p22;q22) translocation in acute myeloid leukemia. Gene Chromosome Canc 2011, 50: 228–238.
Foster N, Paulsson K, Sales M, Cunningham J, Groves M, O’Connor N, Begum S, Stubbs T, McMullan DJ, Griffiths M, Pratt N, Tauro S: Molecular characterisation of a recurrent, semi-cryptic RUNX1 translocation t(7;21) in myelodysplastic syndrome and acute myeloid leukaemia. Br J Haematol 2010, 148: 938–943. 10.1111/j.1365-2141.2009.08039.x
Stankiewicz P, Shaw CJ, Withers M, Inoue K, Lupski JR: Serial segmental duplications during primate evolution result in complex human genome architecture. Genome Res 2004, 14: 2209–2220. 10.1101/gr.2746604
Grimwade D, Howe K, Langabeer S, Davies L, Oliver F, Walker H, Swirsky D, Wheatley K, Goldstone A, Burnett A, Solomon E: Establishing the presence of the t(15;17) in suspected acute promyelocytic leukaemia: cytogenetic, molecular and PML immunofluorescence assessment of patients entered into the M.R.C. ATRA trial. M.R.C. Adult Leukaemia Working Party. Br J Haematol 1996, 94: 557–573.
Nacheva E, Holloway T, Brown K, Bloxham D, Green AR: Philadelphia-negative chronic myeloid leukaemia: detection by FISH of BCR-ABL fusion gene localized either to chromosome 9 or chromosome 22. Br J Haematol 1994, 87: 409–412. 10.1111/j.1365-2141.1994.tb04933.x
Specchia G, Albano F, Anelli L, Zagaria A, Liso A, La Starza R, Mancini M, Sebastio L, Giugliano E, Saglio G, Liso V, Rocchi M: Insertions generating the 5΄RUNX1/3΄CBFA2T1 gene in acute myeloid leukemia cases show variable breakpoints. Gene Chromosome Canc 2004, 41: 86–91. 10.1002/gcc.20061
Kim YK, Kim YS, Yoo KJ, Lee HJ, Lee DR, Yeo CY, Baek KH: The expression of Usp42 during embryogenesis and spermatogenesis in mouse. Gene Expr Patterns 2007, 7: 143–148. 10.1016/j.modgep.2006.06.006
Hock AK, Vigneron AM, Carter S, Ludwig RL, Vousden KH: Regulation of p53 stability and function by the deubiquitinating enzyme USP42. EMBO J 2011, 30: 4921–4930. 10.1038/emboj.2011.419
Zhang L, Lu HH, Chung WY, Yang J, Li WH: Patterns of segmental duplication in the human genome. Mol Biol Evol 2005, 22: 135–141. 10.1093/molbev/msh262
Bailey JA, Eichler EE: Primate segmental duplications: crucibles of evolution, and disease. Nat Rev Genet 2006, 7: 898. 10.1038/nrg1895
Mefford HC, Eichler EE: Duplication hotspots, rare genomic disorders, and common disease. Curr Opin Genet Dev 2009, 19: 196–204. 10.1016/j.gde.2009.04.003
Lupski JR: Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet 1998, 14: 417–422. 10.1016/S0168-9525(98)01555-8
Barbouti A, Stankiewicz P, Nusbaum C, Cuomo C, Cook A, Höglund M, Johansson B, Hagemeijer A, Park SS, Mitelman F, Lupski JR, Fioretos T: The breakpoint region of the most common isochromosome, i(17q), in human neoplasia is characterized by a complex genomic architecture with large, palindromic, low-copy repeats. Am J Hum Genet 2004, 74: 1–10. 10.1086/380648
Albano F, Anelli L, Zagaria A, Coccaro N, D’Addabbo P, Liso V, Rocchi M, Specchia G: Genomic segmental duplications on the basis of the t(9;22) rearrangement in chronic myeloid leukemia. Oncogene 2010, 29: 2509–2516. 10.1038/onc.2009.524
Ebert G, Steininger A, Weißmann R, Boldt V, Lind-Thomsen A, Grune J, Badelt S, Heßler M, Peiser M, Hitzler M, Jensen LR, Müller I, Hu H, Arndt PF, Kuss AW, Tebel K, Ullmann R: Distribution of segmental duplications in the context of higher order chromatin organisation of human chromosome 7. BMC Genomics 2014, 15: 537. 10.1186/1471-2164-15-537
An International System for Human Cytogenetic Nomenclature. 2013.
Lichter P, Tang Chang CJ, Call K, Hermanson G, Evans GA, Housman D, Ward DC: High-resolution mapping of human chromosome 11 by in situ hybridization with cosmid clones. Science 1990, 247: 64–69. 10.1126/science.2294592
Acknowledgements
The authors would like to thank Ms. MVC Pragnell, B.A. for language revision of the manuscript.
This work was supported by “Associazione Italiana contro le Leucemie (AIL)-BARI”.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Zagaria, A., Anelli, L., Coccaro, N. et al. 5‘RUNX1-3’USP42 chimeric gene in acute myeloid leukemia can occur through an insertion mechanism rather than translocation and may be mediated by genomic segmental duplications. Mol Cytogenet 7, 66 (2014). https://doi.org/10.1186/s13039-014-0066-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13039-014-0066-7