Abstract
Background
DNA barcoding can be used to authenticate Ganoderma species for safe use. This study aims to identify commercial products containing Ganoderma using DNA barcoding.
Methods
We used 63 internal transcribed spacer (ITS) 2 sequences of Ganoderma species from 33 newly-sequenced wild samples, crude drugs, mycelia, spores, and authentic extracts and spore oils collected from various locations and markets, as well as 30 sequences from GenBank. Sequences were assembled and aligned using CodonCode Aligner V3.71. Intra- and inter-specific distances were estimated by MEGA 6.0, and phylogeny reconstruction was based on the K2P model. SNP(s) and RNA secondary structure of ITS2 were analyzed and compared among closely related Ganoderma species.
Results
G. lucidum cultivated in China was different from those cultivated in Europe. “Chizhi” (G. lucidum) and “Zizhi” (G. sinense) were clustered into two clades that were separated from the other Ganoderma species. The fruiting bodies and commercial products of G. lucidum and G. sinense were successfully distinguished from those of other Ganoderma species by comparing the ITS2 sequences and RNA secondary structures.
Conclusion
The DNA barcoding method is applicable to the authentication of commercial products containing Ganoderma species.
Similar content being viewed by others
Background
Ganoderma (Lingzhi) is widely used in health products for its anti-tumor, anti-aging, anti-bacterial, immune system-enhancing, and anti-hypertension activities [1–5]. Lingzhi and its derivative products have a world trade value of approximately four billion US dollars [6]. Lingzhi products are popular in the market because of their high demand and potential profits.
There are approximately 76 Ganoderma species in China [7], but only approximately 20 of the species are used for medical purposes [8]. Moreover, only Ganoderma lucidum (Leyss. ex Fr.) Karst., 1881 (Chizhi) and G. sinense Zhao, Xu et Zhang, 1979 (Zizhi) are officially described in the Chinese Pharmacopoeia [9], and they are the most common types of Lingzhi on the market. They are difficult to distinguish because of the intra-species diversity of morphological features [8].
G. lucidum was collected from the UK. Karsten (1881) established the genus Ganoderma based on G. lucidum [10], which was reported in China in 1934 and was first successfully artificially cultivated in 1969 [11]. Cao et al. [11] proposed a new species name, G. lingzhi in 2012 for the Lingzhi that is distributed in East Asia. However, Wang et al. [12] determined that the widely cultivated G. lucidum in China was, in fact, G. sichuanense based on morphological and molecular evidence. Although they provided descriptions for the Lingzhi species in China, they did not obtain sequences from type specimens of G. sichuanense. The genome sequence of G. lucidum in China was first published by our research group [13]. The taxonomy of Lingzhi in China is still under dispute.
The internal transcribed spacer (ITS) region was proposed as a global DNA barcode sequence for identification of fungi at the fourth International Barcode of Life Conference [14]. Chen et al. [15] proposed the nuclear ribosomal DNA second internal transcribed spacer (ITS2) locus as a novel universal DNA barcode to identify herbs based on 6600 samples that represented 4800 species. Han et al. [16] compared the ITS and ITS2 regions and found that ITS2 was more suitable for species identification because of its short length and high efficiency for PCR amplification of this region. Moreover, the sequences and secondary structures of ITS2 could be considered as molecular morphological characteristics for species identification [17]. Considering DNA degradation in Lingzhi products, especially Lingzhi extracts and spore oil, the shorter sequence of ITS2 would likely provide a higher amplification and identification efficiency.
This study aims to authenticate commercial products containing Ganoderma using the DNA barcoding method.
Methods
Sample collection and data acquisition
Sixty-three specimens belonging to 11 Ganoderma species were analyzed. Specimens included 33 samples of commercially cultivated fruiting bodies, strains, slices, spore powders, extracts and spore oils collected in this study, and 30 sequences obtained from GenBank (Table 1). Twenty-six samples of G. lucidum, five strains of G. sinense, and two samples of G. resinaceum were collected in this study. Voucher samples were deposited in the herbarium of the Institute of Medicinal Development at the Chinese Academy of Medicinal Science, Beijing, China. Other published Ganoderma ITS2 sequences were downloaded from GenBank and were also analyzed for their ability to identify species in this study. We screened 348 ITS sequences named G. lucidum (or G. lingzhi). Sequences that met the following criteria were selected: (1) the sequences had already been published; (2) the sequences had complete ITS2 regions; and (3) sequences with original samples that were not from East Asia and Europe or if the original location of the sample was unknown, would be abandoned. The original samples of European G. lucidum for ITS2 sequences that we selected were identified based on morphological features by Yun Cao during previous Ganoderma research [11] and were stored in the Mycological Herbarium, Institute of Microbiology, Chinese Academy of Sciences (HMAS).
DNA extraction, PCR amplification, cloning and sequencing
Specimens were divided into three groups. One group included fruiting bodies, slices, spore powders and extracts. Samples of approximately 30 mg were needed and were ground into powder using a Retsch MM400 (Retsch Co., Germany). Strains (50 mg) were homogenized in liquid nitrogen. Spore oil (300 µL) was first centrifuged at 10,625×g for 10 min using a Sigma 1-14K (Sigma Co., Germany), and the pellet was used for DNA extraction. Total genomic DNA was subsequently extracted using the Plant Genomic DNA kit (Tiangen Biotech Co., China) following the recommended protocol. One pair of primers, 156 (5′-AACCATCGAGTCTTTGAACGC-3′) and 157 (5′-CCTTGTAAGTTTCTTTTCCTCC-3′), were designed for PCR amplification of the ITS2 region of Ganoderma. PCR was performed in 25-µL reaction mixtures, containing 12.5 µL of 2 × PCR buffer (Aidlab Biotechnologies Co., China), 1 µL of each PCR primer (2.5 µM), and 2 µL of DNA extract, and the total volume was adjusted to 25 µL with sterile deionized water. PCR amplification was conducted according to the following procedure: 94 °C for 5 min, 40 cycles of 94 °C for 30 s, 50 °C for 30 s, and 72 °C for 1 min, and a final extension at 72 °C for 10 min. PCR products were analyzed by electrophoresis in a 1 % agarose gel. The PCR products were purified using the PCR Purification Kit (Tiangen Biotech Co., China) and sequenced bidirectionally using a ABI 3730XL sequencer (Applied Biosystems Co., USA) based on the Sanger sequencing method at the Genome Center, Chinese Academy of Agricultural Sciences.
Phylogenetic analysis
The sequences were edited and assembled manually using CodonCode Aligner V3.71 (CodonCode Co., USA). The new sequences obtained in this study were deposited in GenBank. ITS sequences from GenBank were annotated using the Hidden Markov model (HMM) [18] to obtain the ITS2 sequences. All ITS2 sequences were included in the phylogenetic analysis by MEGA 6.0 [19]. All of the sequences were aligned using the MUSCLE method [20]. A neighbor-joining (NJ) [21] tree was constructed with the following parameters: the bootstrap method was conducted with 1000 replicates, the substitution model was Kimura-2-parameter (K2P) [22], and gaps were treated as missing data (complete deletion). Maximum parsimony (MP) [23] trees were constructed with the following parameters: the bootstrap method was conducted with 1000 replicates, the MP search method was subtree pruning and regrafting [24], the number of initial trees was ten (random addition), and gaps were treated as missing data (complete deletion). Sequence divergence was also calculated using the K2P model, and gaps were treated as missing data. Tomophagus colossus was selected as the outgroup. The secondary structure of ITS2 was predicted at the ITS2 database website (http://www.its2.bioapps.biozentrum.uni-wuerzburg.de/) [25].
Results
ITS2 sequence analysis and intra- and inter-species variations
The PCR product sizes for the ITS2 region ranged from 469 to 566 bp. The length of ITS2 was 218 bp after deletion of the 5.8S and 28S rDNAs and alignment using the MUSCLE method. The average G-C and A-T contents of the ITS2 region were 49.4 and 50.6 %, respectively. The aligned ITS2 rDNA sequences are shown in Fig. 1. The 26 newly collected samples of G. lucidum from China had seven intraspecific variable sites (Fig. 1), and 10 of these 26 samples had identical sequences. The ITS2 regions of G. sinense resulted in ambiguous sequences with direct sequencing of the PCR products; thus, a cloning method was used.
Multiple sequence alignment of 63 ITS2 sequences. The sequences were aligned using MUSCLE. Fifty-three variable sites in the 218-bp sequence alignment of the ITS2 were extracted and presented. The specimen names are shown in the left side of the alignment. Gaps are indicated with dashes, and identical sites are indicated with dots
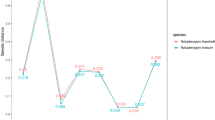
Nucleotide analysis of the ITS2 region could provide more information about inter-and intra-species divergences. The average intraspecific genetic distances calculated by the Kimura-2-parameter model [22] were 0.007 for G. lucidum from East Asia. No variable sites were detected among the ITS2 regions of nine G. sinense samples collected from Shandong, Hubei and Hainan. The interspecific diversities ranged from 0.035 to 0.047 between G. lucidum from Europe and G. lucidum from East Asia, from 0.097 to 0.111 between G. lucidum from East Asia and G. sinense, and from 0.035 to 0.123 between G. lucidum from East Asia and the other species examined. In this study, the intra-species distances of the Ganoderma species were lower than the inter-species distances except for in G. sichuanense and G. weberianum (Fig. 2).
SNP-based molecular barcodes have been used for identification studies in closely related species [26]. There were seven stable SNPs existing between G. lucidum from East Asia and G. lucidum from Europe, including sites of deletion/insertion, and 25 stable SNPs between G. lucidum from East Asia and G. sinense including four sites of deletion/insertion (Fig. 1). At positions 23, 76, 108 and 196 bp, all G. lucidum samples from Europe contained C, G, A, and G, respectively. Meanwhile, all G. lucidum samples from East Asia contained T, A, G, and T, respectively.
Phylogenetic analysis
Species of the genus Ganoderma, including G. lucidum (from Europe), G. sinense, G. applanatum, G. fornicatuma, G. multipileum, G. resinaceum, G. sichuanense, G. weberianum, G. tenue, and G. tropicum, which are closely related to G. lucidum (from East Asia), were used to study the relationships between Lingzhi species. Sixty-three ITS2 sequences were analyzed. A total of 218 characters were included for phylogenetic analysis, of which 61 were variable and 53 were parsimony informative characters. The consistency index was 0.6914, the retention index was 0.9324, and the composite index was 0.6614 for all sites and parsimony-informative sites (in parentheses).
The topologies of the NJ and MP trees (Fig. 3) were similar. The high level of nucleotide substitution in the ITS2 rDNA resulted in six clades. Although most sequences of either G. lucidum or G. sinense had identical ITS2 sequences, G. lucidum from Europe did not fit into these clades. In the phylogenetic trees, Group 1 consists of collections from G. lucidum from East Asia; 28 sequences of G. lucidum from China and Korea and five sequences of G. lucidum from Europe were clustered into two distinct clades, which were separate from the other species with high bootstrap support values. The three unknown samples from Taiwan (SN04MT24, SN04MT25, and SN04MT26) clustered with G. lucidum from East Asia, and the Dai12573 strains of G. lingzhi were within the same group. These data strongly indicated that G. lucidum from East Asia was not the same species as G. lucidum from Europe, and that G. lucidum could be misnamed in Asia. Sequences of G. sinense and G. japonicum formed a high-support value clade (100 %). G. sinense Zhao, Xu et Zhang is a new species that Zhao et al. established in 1979 [27] to eliminate confusion with G. japonicum (Fr.) Lloyd. Our results confirmed that the two species should be synonymous because the ITS2 sequence of G. sinense was identical to that of G. japonicum.
Phylogenetic tree based on the ITS2 region using the NJ method. Strict consensus phylogenetic trees constructed using MP (a) and NJ (b) methods based on the ITS2 sequences of 63 taxa of Ganoderma. Bootstrap values are shown above the branches. Based on the trees, the taxa can be divided into six clades
Group 5 consisted of two subgroups (100 % bootstrapping): subgroup 5.1 included G. sichuanense, G. weberianum and G. tenue, and subgroup 5.2 consisted of four G. resinaceum samples. G. sichuanense, G. weberianum, and G. tenue were grouped into one well-supported clade (94.3 %), but the relationships among these species require further study. Two samples of G. multipileum from China clustered together and formed a clade with G. lucidum from Asia. G. multipileum, a species for which there was a holotype specimen from Taiwan, was suggested as the correct name for the tropical Ganoderma samples and showed a close relationship with G. lucidum from East Asia.
Efficiency of species identification
BLAST1 was used to further evaluate the efficiency of ITS2. The barcode sequences obtained in this study were used to build corresponding reference sequence libraries as described previously [28]. The results showed that ITS2 successfully identified 100 % of the commercial Lingzhi products collected in this study.
The ITS2 sequence-structure provided the most accurate phylogenetic analysis [29], and ITS2 sequence-structure information was correlated with the biological species concept [30]. Thus, the RNA secondary structures of ITS2 were analyzed to differentiate the species of G. lucidum. The three closely-related species have similar secondary structures of ITS2 sequences. Stem-loops I, II, and III were conserved, whereas stem-loop IV of the three species varied. The three species could be identified directly based on the RNA secondary structure of ITS2 (Fig. 4).
Discussion
G. lucidum is one of the most economically important species of fungi; thus the stability of its taxonomy is highly important. Different researchers have different views on the scientific binomial of “Chizhi” [11, 12, 31–33]. In China, Lingzhi has long been misnamed G. lucidum. We identified commercial Lingzhi products based on the ITS2 rDNA marker, and our phylogenetic analysis clearly indicated that Chizhi, or G. lucidum, from East Asia, is not the same species as G. lucidum from Europe. The nucleotide divergence among G. lucidum from Europe and G. lucidum from East Asia, as well as the high bootstrapping support, indicated that they were different species. Analysis of RNA secondary structure further supported these results.
Identifying Ganoderma products (such as spore oil and extracts) according to morphological characteristics alone is difficult. The triterpenes of G. lucidum and G. sinense show significant differences in terms of types and content, and a distinction should be made between the medical uses of the two species [34–38]. The use of systematic methods for species identification and classification would be useful. The evolutionary context of the related species should be studied first to identify the biological species. An ideal barcode sequence should possess high inter-species divergence but low intra-species divergence to readily identify different species. ITS2 has a wider taxonomic coverage than was previously assumed because of the high sequence variability and conserved core secondary structure [16]. Moreover, ITS2 had comparable power for resolving closely related species, and especially for identifying herbs and specimens that have undergone DNA degradation. The nuclear ribosomal DNA second internal transcribed spacer ITS2 sequence is a double-edged tool for eukaryotic evolutionary comparisons [39], and has been proven useful for diagnostic purposes at the species level [40]. In the present study, we analyzed the ITS2 region of Ganoderma species to accurately identify commercial Lingzhi products. Our results showed that most Ganoderma products, including Chizhi (G. lucidum), Zizhi (G. sinense) and other Ganoderma species, could be successfully identified using ITS2 sequences. Our results also support the suggestion that G. sinense and G. japonicum should be considered synonymous because of their high sequence similarity.
In this study, regardless of the complicated taxonomy of Ganoderma, the sequence-based phylogeny supported the hypothesis that G. lucidum species originating in Europe and East Asia are not the same species.
Conclusion
The DNA barcoding method is applicable to the authentication of commercial products containing Ganoderma species.
References
Jong SC, Birmingham JM (1992) Medicinal benefits of the mushroom Ganoderma. Adv Appl Microbiol 37:101–34
Lin ZB (2001) Modern research of Ganoderma, 2nd edn. Beijing Medical University Press, Beijing
Tan BKH, Vanitha J (2004) Immunomodulatory and antimicrobial effects of some traditional Chinese medicinal herbs: a review. Curr Med Chem 11(11):1423–1430
Paterson RRM (2006) Ganoderma–A therapeutic fungal biofactory. Phytochemistry 67(18):1985–2001
Boh B, Berovic M, Zhang J, Zhi-Bin L (2007) Ganoderma lucidum and its pharmaceutically active compounds. In: El-Gewely MR (ed) Biotechnology annual review. Elsevier, The Netherlands, pp 265–301
Perumal K (ed) (2009) Indigenous Technology on Organic Cultivation of Reishi (Ganoderma lucidum) in India. Proceedings of the 5th International Medicinal Mushroom Conference, Nantong, Jiangsu, China
Zhang XQ, Zhao JD (2000) Mycoflora of China Ganodermataceae, vol 18. Science Press, Beijing
Shi FM, Ding ZM, Chen SL, Tong XR (2012) Research Progress on Resource and Identification of Ganoderma. World Sci Technol Modern Tradit Chin Med Mater Med 2:1473–1480. doi:10.3969/j.issn.1674-3849.2012.02.028
China PCoPsRo (ed) (2010) Pharmacopoeia of the People’s Republic of China. China Medical Science Press, Beijing
Karsten PA (1881) Enumeratio Boletinearum et Polyporearum Fennicarum, systemate novo dispositarum. Revue Mycologique Toulouse. 3(9):4
Cao Y, Wu S-H, Dai Y-C (2012) Species clarification of the prize medicinal Ganoderma mushroom “Lingzhi”. Fungal Divers 56(1):49–62
Wang XC, Xi RJ, Li Y, Wang DM, Yao YJ (2012) The species identity of the widely cultivated Ganoderma, ‘G. lucidum’ (Ling-zhi), in China. PLoS One 7(7):e40857
Chen S, Xu J, Liu C, Zhu Y, Nelson DR, Zhou S et al (2012) Genome sequence of the model medicinal mushroom Ganoderma lucidum. Nature Commun 3:913. doi:10.1038/ncomms1923
Zhang Y, Guo LD (2012) Progress of fungal DNA barcode. Mycosystema 31(6):809–820
Chen S, Yao H, Han J, Liu C, Song J, Shi L et al (2010) Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS One 5(1):e8613
Han J, Zhu Y, Chen X, Liao B, Yao H, Song J et al (2013) The short ITS2 sequence serves as an efficient taxonomic sequence tag in comparison with the full-length ITS. BioMed Res Int 2013;2013:741476. doi:10.1155/2013/741476
Grajales A, Aguilar C, Sánchez JA (2007) Phylogenetic reconstruction using secondary structures of Internal Transcribed Spacer 2 (ITS2, rDNA): finding the molecular and morphological gap in Caribbean gorgonian corals. BMC Evol Biol 7(1):90
Keller A, Schleicher T, Schultz J, Muller T, Dandekar T, Wolf M (2009) 5.8S-28S rRNA interaction and HMM-based ITS2 annotation. Gene 430(1–2):50–57
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32(5):1792–1797
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16(2):111–120
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Biol 20(4):406–416
Nei M, Kumar S (2000) Molecular evolution and phylogenetics. Oxford University Press, Oxford
Koetschan C, Förster F, Keller A, Schleicher T, Ruderisch B, Schwarz R et al (2010) The ITS2 database III—sequences and structures for phylogeny. Nucleic Acids Res 38(suppl 1):D275–D279
Chen X, Liao B, Song J, Pang X, Han J, Chen S (2013) A fast SNP identification and analysis of intraspecific variation in the medicinal Panax species based on DNA barcoding. Gene 530(1):39–43
Zhao JD, Xu LW, Zhang XQ (1979) Taxonomic studies on the subfamily Ganodermiodeae of China. Acta Microbiol Sinica 19(3):265–279
Xiang L, Song J, Xin T, Zhu Y, Shi L, Xu X et al (2013) DNA barcoding the commercial Chinese caterpillar fungus. FEMS Microbiol Lett 347(2):156–162
Keller A, Förster F, Müller T, Dandekar T, Schultz J, Wolf M (2010) Including RNA secondary structures improves accuracy and robustness in reconstruction of phylogenetic trees. Biol Direct 5(4):1–12
Müller T, Philippi N, Dandekar T, Schultz J, Wolf M (2007) Distinguishing species. RNA 13(9):1469–1472
Park YJ, Kwon OC, Son ES, Yoon DE, Han W, Yoo YB et al (2012) Taxonomy of Ganoderma lucidum from Korea Based on rDNA and Partial beta-Tubulin Gene Sequence Analysis. Mycobiology 40(1):71–75
Pegler DN, Yao YJ (1996) Oriental species of Ganoderma section Ganoderma. Botany and mycology for the next millenium: collection of scientific articles devoted to the 70th Anniversary of Academician Sytnik KM Kyiv: Kholodny NG Institute of Botany, National Academy of Sciences of Ukraine, pp 336–47
Wang DM, Wu SH, Su CH, Peng JT, Shih YH, Chen LC (2009) Ganoderma multipileum, the correct name for ‘G. lucidum’ in tropical Asia. Bot Stud 50:451–458
Lu J, Qin JZ, Chen P, Chen X, Zhang YZ, Zhao SJ (2012) Quality difference study of six varieties of Ganoderma lucidum with different origins. Front Pharmacol 3:57. doi:10.3389/fphar.2012.00057
Da J, Wu WY, Hou JJ, Long HL, Yao S, Yang Z et al (2012) Comparison of two officinal Chinese pharmacopoeia species of Ganoderma based on chemical research with multiple technologies and chemometrics analysis. J Chromatogr A 1222:59–70
Ding P, Yu QX, Liang YJ, Liang JY, Wang HL (2009) Study on HPTLC chromatographic fingerprint analysis of Ganoderma lucidum and its related species. Chin Pharm J 44(24):1854–1857
Ding P, Huang HB, Qiu JY, Liang YJ, Wang HL (2009) Study on the differences of Ganoderma lucidum and Ganoderma sinense by HPTLC and HPLC fingerprint chromatogram. West Chin J Pharm 4:404–406
Wang XM, Yang M, Guan SH, Liu RX, Xia JM, Bi KS et al (2006) Quantitative determination of six major triterpenoids in Ganoderma lucidum and related species by high performance liquid chromatography. J Pharm Biomed Anal 41(3):838–844. doi:10.1016/j.jpba.2006.01.053
Coleman AW (2003) ITS2 is a double-edged tool for eukaryote evolutionary comparisons. Trends Genet 19(7):370–375
Prasad PK, Tandon V, Biswal DK, Goswami LM, Chatterjee A (2009) Use of sequence motifs as barcodes and secondary structures of Internal Transcribed spacer 2 (ITS2, rDNA) for identification of the Indian liver fluke, Fasciola (Trematoda: Fasciolidae). Bioinformation 3(7):314
Park YJ, Kwon OC, Son ES, Yoon DE, Han W, Nam JY et al (2012) Genetic diversity analysis of Ganoderma species and development of a specific marker for identification of medicinal mushroom Ganoderma lucidum. Afr J Microbiol Res 6(25):5417–5425
Su CL, Tang CH, Zhang JS, Chen MJ, Pan YJ (2007) The phylogenetic relationship of cultivated isolates of Ganoderma in China inferred from nuclear ribosomal DNA ITS sequences. Acta Microbiol Sinica 1:11–16
Wang DM, Yao YJ (2005) Intrastrain internal transcribed spacer heterogeneity in Ganoderma species. Can J Microbiol 51(2):113–121
Guglielmo F, Gonthier P, Garbelotto M, Nicolotti G (2008) A, PCR-based method for the identification of important wood rotting fungal taxa within Ganoderma, Inonotus s.l. and Phellinus s.l. FEMS Microbiol Lett 282(2):228–237
Moncalvo JM, Wang HF, Hseu RS (1995) Gene phylogeny of the Ganoderma lucidum complex based on ribosomal DNA sequences. Comparison with traditional taxonomic characters. Mycol Res 99(12):1489–1499
Le X, Le Nguyen Q, Pham N, Duong V, Dentinger BM, Moncalvo J-M (2012) Tomophagus cattienensis sp. nov., a new Ganodermataceae species from Vietnam: Evidence from morphology and ITS DNA barcodes. Mycol Progress 11(3):775–780
Authors’ contributions
JPH conceived and designed the study. BSL, XCC, LLW, and WJJ performed data analyses. BSL and JPH wrote the manuscript. America Journal experts had revised the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This research was supported by Grants from the Major Scientific and Technological Special Project for “Significant New Drugs Creation” (No. 2014ZX09304307001) and the National Natural Science Foundation of China (Grant No. 81473303). We thank our colleagues who helped with sample collection, identification, laboratory work and manuscript preparation, including Professors Yulin Lin, Caixiang Xie, and countless others.
Compliance with ethical guidelines
Competing interests The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Liao, B., Chen, X., Han, J. et al. Identification of commercial Ganoderma (Lingzhi) species by ITS2 sequences. Chin Med 10, 22 (2015). https://doi.org/10.1186/s13020-015-0056-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13020-015-0056-7