Abstract
Background
Estrogen receptor beta (ERβ) was considered as a tumor-inhibiting factor in estrogen-sensitive malignant tumors. In this study, we intended to investigate whether ERβ was involved in inducing autophagy in osteosarcoma.
Methods
This is an experimental study. The associations between ERβ and autophagy were detected in osteosarcoma U2-OS cells which were treated with E2, E2 + 2,3-Bis (4-hydroxyphenyl) propionitrile (DPN, ERβ agonists), E2 + DPN + water, E2 + DPN + 3-Methyladenine (3-MA, autophagy inhibitor), respectively. Cell viability and death were detected using cell counting kit 8 assay and flow cytometry, respectively. In addition, the expression of autophagy marker LC3II/I, sequestosome 1 (P62), mammalian target of rapamycin (mTOR), and phosphorylated-mTOR (p-mTOR) was determined by reverse transcription quantitative polymerase chain reaction (RT-qPCR) and Western blotting.
Results
Cell viability was significantly decreased with DPN treatment, while was reversed with 3-MA treatment. DPN treatment decreased living cells proportion and increased cell apoptosis proportion, while 3-MA treatment reversed those changes. However, there were significant differences between the E2 group and the E2 + DPN + 3-MA group for the living cell proportion and cell apoptosis proportion, suggesting apoptosis and autophagy all were induced. In addition, DPN treatment upregulated the LC3II/I expression level and downregulated P62 and mTOR (mRNA level) and p-mTOR (protein level) expression levels.
Conclusion
ERβ inhibited the cell viability and mediated cell death by inducing apoptosis and autophagy in osteosarcoma. ERβ-induced autophagy in osteosarcoma was associated with downregulating the P62 expression level and inhibiting mTOR activation.
Similar content being viewed by others
Background
Osteosarcoma is the most common primary malignant tumor in bone tissue that mainly occurs in adolescents and the elderly, and it is characterized by proliferating tumor cells that directly form bone or bone-like tissue [1]. The incidence of osteosarcoma in males is higher than that in females and commonly emerges in the distal femur, proximal tibia, and proximal humerus [2]. The current treatment of osteosarcoma is mainly neoadjuvant chemotherapy and surgery, and the 5-year overall survival rate has increased to about 70 to 80% because of the introduction of neoadjuvant chemotherapy and radiotherapy [3, 4]. However, the prognosis of patients with osteosarcoma is poor, and metastasis is the most important prognostic factor, followed by chemotherapy response, tumor characteristics, relapse, and so on [4]. The overall survival of the patients with metastasis or recurrence is less than 20% in spite of chemotherapeutic drugs [5]. In addition, drug-resistance and severe side effects, including cardiotoxicity and nephrotoxicity, are the adverse factors of chemotherapy [6]. Therefore, it is necessary to further study the inner mechanism of the development and progression of osteosarcoma to provide a theoretical basis for clinical treatment.
Estrogen receptors (ER) belong to the nuclear receptor superfamily, and these ligand-dependent receptors respond to the presence of estrogen [7]. The changes in the ER expression level or tissue distribution are observed in multiple cancers [8]. The two subtypes, ERα and ERβ, are highly homologous in structure, while being different in tissue distribution and the biological effects under binding to ligand [9]. Reportedly, ERβ serves an important role in autophagy. For instance, Pierdominici et al. reported that ERβ activation can repress the progression of Hodgkin lymphoma by inducing autophagy [10]. Nevertheless, there are no reports focusing on whether ERβ is implicated in mediating autophagy in osteosarcoma. In our previous study, we found that ERβ mediated the proliferation, migration, and invasion of osteosarcoma cells by the regulation of the integrin, Bcl-2, and PI3K/Akt signal pathways [11]. Hence, we intended to investigate whether ERβ is involved in inducing autophagy in osteosarcoma. 2,3-Bis (4-hydroxyphenyl) propionitrile (DPN) was a selective agonist of ERβ and it had been used to examine the role of ERβ in studies [12, 13], while 3-methyladenine (3-MA) was an inhibitor of autophagy [14]. In our study, DPN and 3-MA were selected to examine the association among ERβ and autophagy on osteosarcoma cell lines U2-OS.
Methods
Cell culture and treatment
The osteosarcoma cell lines U2-OS were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China), and cultured in RPMI-1640 medium (catalog no. C22400500BT, Gibco) supplemented with 10% fetal bovine serum (FBS, catalog no. 10099-141, Gibco) and 1% penicillin-streptomycin (catalog no. BS734, Sangon Biotech) in an incubator with 5% CO2 at 37 °C. The U2-OS cells under logarithmic growth phase were used in subsequent experiments. There were four groups in this study.
Group A: NC + 1 uM E2 (estradiol; catalog no. E808987, Macklin);
Group B: NC + 1 uM E2 + 100 uM DPN (catalog no. H5915, Sigma);
Group C: NC + 1 uM E2 + 100 uM DPN + water (solvent);
Group D: NC + 1 uM E2 + 100uM DPN + 1 mM 3-MA (catalog no. HY-19312, MCE).
The concentrations of E2, DPN, and 3-MA were determined by half-maximal inhibitory concentration (IC50).
Cell counting kit-8 assay
Cells suspension was prepared at 5 × 104 cells/ml and placed at 100 μl/well in 96-well plate overnight at 37 °C. After 48 h of treatments, the original medium was replaced with serum-free medium containing 10% of 5 mg/ml cell counting kit-8 (CCK-8) solution (catalog no. C0039, Beyotime) at 100 μl/well in 96-well plate, and incubated in dark for 2 h. Then, the light absorption value of each well at 450 nm was measured using a microplate reader (Infinite M100 PRO, TECAN).
Apoptosis analysis
Cells were placed in a six-well plate at a density of 3 × 105 cells/well overnight, and three duplicates were set for each group. After 48 h of treatments, apoptosis was determined by flow cytometric analysis. In short, cells were resuspended in 100 μL 1× binding buffer coupled with 5 μl FITC-Annexin-V (catalog no. 40301ES50, BD) and 5 μl of 50 μg/mL propidium iodide (Sigma), and incubated in the dark for 2 h at room temperature. Then, cell apoptosis was measured using FACS Calibur flow cytometer (BD) after adding 400 μL of 1x binding buffer.
Reverse transcription quantitative polymerase chain reaction
Total RNA was extracted using TRIzol reagent (catalog no. 15596026, Invitrogen; Thermo) in the light of instructions. The quality and purity of total RNA were measured by a microplate reader (Infinite M100 PRO, TECAN). The total RNA was reverse transcribed using PrimeScript™ RT Master Mix (catalog no. RR036A, Takara). Then, RT-qPCR was performed using Power SYBR Green PCR Master Mix (catalog no. A25742, Thermo). The thermal cycling conditions were as follows: 50 °C for 3 min, 95 °C for 3 min, followed by 40 cycles at 95 °C for 10 s and 60 °C for 30 s. Melt curve was analyzed from 60 to 95 °C at an increment rate of 0.5 °C/10 s. The relative expression of the genes was calculated by the 2−ΔΔCt method [15]. The primer sequences for the genes were as follows: microtubule-associated protein 1 Light Chain 3 (LC3), LC3II/I, forward 5′-AACATGAGCGAGTTGGTCAAG-3′; LC3II/I, reverse 5′-GCTCGTAGATGTCCGCGAT-3′; Sequestosome 1 (P62), forward 5′-GGAACAGCGACTCTTGCTTC-3′; reverse 5′-GGTGCTCGATATGGCATTAGTG-3′; mammalian target of rapamycin (mTOR), forward 5′-TCCGAGAGATGAGTCAAGAGG-3′; reverse 5′-CACCTTCCACTCCTATGAGGC-3′;GAPDH, forward 5′-TGACAACTTTGGTATCGTGGAAGG-3′; GAPDH, reverse 5′-AGGCAGGGATGATGTTCTGGAGAG-3′.
Western blotting
Cells were lysed in RIPA lysis buffer (catalog no. P0013B, Beyotime) coupled with PMSF (catalog no. ST506, Beyotime) at a final concentration of 1 mM, and the protein concentration was determined using a BCA Protein Assay Kit (catalog no. 23227, Thermo). Protein (20 μg) from each sample was separated by 10% SDS-polyacrylamide gel and transferred onto polyvinylidene difluoride (PVDF) membranes (catalog no. IPVH00010, Millipore). Five percent of skim milk (catalog no. 232100, BD) was used to block the nonspecific binding for 1–2 h at 37 °C and then incubated with anti-LC3I/II (1:1000, catalog no. WL01506, Wanleibio), anti-p62 (1:1000, catalog no. 5114S, CST), anti-p65 (1:1000, catalog no. 1074S, CST), anti-P mTOR (1:1000, catalog no. 5536S, CST), anti-mTOR (1:1000, catalog no. 20657-1-AP, Proteintech), anti-β actin (1:1000, catalog no. ab8226, abcam) primary antibody at 4 °C overnight. The membrane was washed with PBST buffer and then incubated with HRP secondary antibodies (1:10000, catalog no.111-035-045, Jackson) for 2 h at 37 °C. After washing with PBST buffer six times, the membrane was exposed with enhanced chemiluminescence (ECL) reagents (catalog no. SB-WB012, Share-bio) and the Millipore ECL system, and then gray scanning was performed using TanonImage (4600, Tanon).
Autophagic flux assay
Cells were placed in a six-well plate at a density of 2 × 105 cells/well, and pLV-GFP-LC3 lentivirus with multiplicity of infection (MOI) of150 and 6 μg/ml of polybrene (catalog no. 107689, sigma) were added. Then, cells were transferred to a 10-cm dish, and puromycin (catalog no. P8833, Ssigma) was added with a final concentration of 2 μg/ml for 72 h when the cell density reached 60%. Finally, the cells transfected with pLV-GFP-LC3 lentivirus were treated with different drugs. The expression of green fluorescence was detected using fluorescence microscopy (TCS SP5, Leica) after 48-h treatments.
Statistical analysis
Statistical analysis was performed using SPSS 22.0 software package (SPSS Inc., Chicago, IL). All experiments were performed in triplicate, and the results were presented with mean value ± standard deviation (SD). The Shapiro-Wilk was used to test the normality of the distribution. For the data presenting a normal distribution, Student’s t test (two groups) and one-way ANOVA (more than two groups) were used to compare the results among different groups. The Wilcoxon rank-sum test was used for nonnormally distributed data. P < 0.05 was selected to show the significant difference.
Results
ERβ inhibited the viability of U2-OS cells
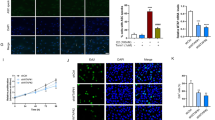
The cells viability were inhibited with DPN treatment compared with that in control (E2 vs. E2 + DPN; E2 vs. E2 + DPN + water; P < 0.001), while the inhibited cells viability were reversed with 3-MA treatment (E2 + DPN + 3-MA vs. E2 + DPN; E2 + DPN + 3-MA vs. E2 + DPN + water; P < 0.001), indicating that ERβ could inhibit the osteosarcoma cells viability probably through inducing autophagy (Fig. 1).
ERβ mediates U2-OS cell death by inducing apoptosis and autophagy
The proportion of living cells were decreased with DPN treatment compared with the control (E2 vs. E2 + DPN; E2 vs. E2 + DPN + water; P < 0.001). Meanwhile, the proportion of living cells were also decreased with 3-MA treatment compared with control (E2 vs. E2 + DPN + 3-MA, P < 0.05), while it was increased compared with DPN treatment (E2 + DPN + 3-MA vs. E2 + DPN; E2 + DPN + 3-MA vs. E2 + DPN + water; P < 0.001). In addition, cell apoptosis was induced with DPN treatment compared with control (E2 vs. E2 + DPN; E2 vs. E2 + DPN + water; P < 0.001). The proportion of apoptotic cells were also increased with 3-MA treatment compared with control (E2 vs. E2 + DPN + 3-MA, P < 0.01), while it was decreased compared with DPN treatment (E2 + DPN + 3-MA vs. E2 + DPN; E2 + DPN + 3-MA vs. E2 + DPN + water; P < 0.001). Those findings suggested that ERβ could mediate cell death in osteosarcoma by inducing apoptosis and autophagy (Fig. 2).
Moreover, the GFP-LC3 fusion protein was dispersed in the cytoplasm in control (E2 group), while there were many green fluorescence spots with DPN treatment (E2 + DPN and E2 + DPN + water group), that were autophagosomes. However, autophagosomes were decreased in E2 + DPN + 3-MA group. The results of autophagic flux assay revealed that ERβ induced autophagy in osteosarcoma cells (Fig. 3).
The expression of LC3-II/I, P62, and mTOR at mRNA and protein levels
To demonstrate the mechanisms of ERβ-induced autophagy in U2-OS cells, the expression of LC3-II/I, P62, and mTOR were determined (Fig. 4). LC3 was an autophagy marker, and LC3-I conversion to LC3-II occurred during autophagy. The expression of LC3-II/I was increased with DPN treatment compared with control (E2 vs. E2 + DPN; E2 vs. E2 + DPN + water; P < 0.01 at mRNA level, P < 0.001 at protein level), while the increased LC3-II/I expression was reversed with 3-MA treatment (E2 + DPN + 3-MA vs. E2 + DPN; E2 + DPN + 3-MA vs. E2 + DPN + water; P < 0.01 at mRNA level, P < 0.001 at protein level).
The expression of LC3-II/I, P62, mTOR, and p-mTOR determined by RT-qPCR and Western blotting. *: E2 vs. E2 + DPN; E2 vs. E2 + DPN + water; E2 vs. E2 + DPN + 3-MA. &: E2 + DPN + 3-MA vs. E2 + DPN; E2 + DPN + 3-MA vs. E2 + DPN + water. * P < 0.05; & P < 0.05; ** P < 0.01; &&P < 0.01; *** P < 0.001; &&&P < 0.001
Nevertheless, the mRNA expression of P62 was decreased with DPN treatment compared with control (E2 vs. E2 + DPN; E2 vs. E2 + DPN + water; P < 0.01), and was reversed with 3-MA treatment (E2 + DPN + 3-MA vs. E2 + DPN; E2 + DPN + 3-MA vs. E2 + DPN + water; P < 0.01). Also, the protein expression of P62 was decreased with DPN treatment compared with control (E2 vs. E2 + DPN, P < 0.01; E2 vs. E2 + DPN + water; P < 0.05), and was reversed with 3-MA treatment (E2 + DPN + 3-MA vs. E2 + DPN; E2 + DPN + 3-MA vs. E2 + DPN + water; P < 0.05). There were no significant differences for the expression of mTOR both at mRNA and protein levels among groups. However, the protein expression of p-mTOR was downregulated with DPN treatment (E2 vs. E2 + DPN, P < 0.01; E2 vs. E2 + DPN + water; P < 0.05), and was reversed with 3-MA treatment (E2 + DPN + 3-MA vs. E2 + DPN; E2 + DPN + 3-MA vs. E2 + DPN + water; P < 0.05). Those findings demonstrated that ERβ induced autophagy by reducing the level of P62 and mTOR.
Discussion
ERs were reported to be involved in the nosogenesis and tumor progression of many cancer, such as prostatic cancer, breast carcinoma, ovarian cancer, and others [16]. ERβ was found to exert tumor-suppressive effects in cancers, suggesting the therapeutic target role of ERβ [17, 18]. For example, a study suggested that Erβ served as a tumor suppressor in prostate cancer by inhibiting the expression and activity of androgen receptor [18]. In addition, ERβ agonists had been selected to prevent and treat in several cancers [19,20,21]. Wei et al. reported that the ERβ-mediated cyclin D1 degradation could repress the proliferation of colon cancer cells by autophagy [22]. In addition, a study had revealed that ERβ overexpression served a neuroprotective role by the interaction with ATG7 and the activation of autophagy—lysosomal [23]. In the current study, we found that the ERβ agonist (DPN) inhibited the osteosarcoma cell viability and mediated the osteosarcoma cell death by inducing apoptosis and autophagy. This was consistent with the above views.
Autophagy was considered as a type II programmed cell death associated with the regulation of intracellular homeostasis [24]. Autophagy referred to a lysosomal degradation process starting at the formation of autophagosomes, and it was involved in multiple physiological processes, such as cell survival, differentiation and death, and pathological processes [24, 25]. Changes of autophagy activity are associated with the development and progression of malignant tumors, and the role of autophagy in anti-tumor therapy is still debatable [25]. LC3 was microtubule-associated protein which was widely present in tissues and cells of mammal [26]. LC3 ubiquitin-like modifications were implicated in the formation of autophagosomes [26]. LC3-I was converted to LC3-II which was transposed to the autophagosome membrane during autophagy, and LC3-II had been considered as an autophagosomal marker [27]. This conversion was also observed in our study.
P62, also named sequestosome 1 or ubiquitin-binding protein P62, was the first identified autophagy adaptor [28]. p62 is also a signaling hub by interacting with key signaling proteins; studies on p62 in cell signaling and autophagy would be extremely important for therapeutic exploitation of such an adaptor protein [28]. In addition, it was reported that P62 could be directly bound to LC3 through a specific motif, and contributed to the connection of ubiquitinated proteins to autophagy mechanism, so that they could be degraded in the lysosome [27]. Xu et al. demonstrated Sp1 transcription factor (SP1) could increase the expression of p62 via binding to p62 promoter, and SP1 could decrease autophagy flux by the activation of p62, while SP1 deficiency could increase autophagy flux in gastric cancer [29]. Zhang et al. suggested that DEAD box protein 5 could inhibit tumorigenesis in hepatocellular carcinoma through inducing autophagy by interacting with p62/sequestosome 1 [30]. In our study, the expression of P62 was significantly decreased with the DPN treatment, while the p62 expression was increased when treated with 3-MA, suggesting that ERβ activation did induce autophagy. Those results were consistent with the previous reports above.
As far as we know, mTOR was a crucial factor in the regulation of autophagy and served an important role in mediating the balance among cell growth and autophagy in answer to nutriture and other signals. The deregulation of the mTOR signaling pathway had been reported to be involved in various pathological diseases and tumor progression, including osteosarcoma [31, 32]. In osteosarcoma, activation of mTOR signaling (phosphorylation) accelerated cellular metastasis and presented an unfavorable prognosis through the regulation of downstream targets [32]. Kim et al revealed that deoxypodophyllotoxin triggered autophagy in osteosarcoma U2-OS cells which was cytoprotective via repressing phosphoinositide-3-kinase (PI3K) /AKT/mTOR pathway [33]. In addition, it had reported that autophagy and apoptosis were triggered in osteosarcoma MG-63 cells with honokiol treatment, which was associated with increasing the expression of LC3II while decreasing PI3K, phosphorylation-Akt, and p-mTOR expression levels [34]. Similarly, p-mTOR expression was significantly reduced with DPN treatment while increased with 3-MA treatment in our study. From the above, we concluded that ERβ repressed osteosarcoma progression through triggering autophagy which was associated with the expression of P62 and p-mTOR. However, the specific regulatory mechanism had not been clearly explored; for instance, the downstream effectors were need to be investigated.
Conclusion
In a word, ERβ inhibited cell viability and mediated cell death by inducing apoptosis and autophagy in osteosarcoma. ERβ-induced autophagy in osteosarcoma was associated with downregulating the expression of P62 and p-mTOR. The current study revealed that ERβ might be a promising target for the study and the treatment of osteosarcoma.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- 3-MA:
-
3-Methyladenine
- CCK-8:
-
Cell counting kit-8
- ECL:
-
Enhanced chemiluminescence
- ER:
-
Estrogen receptors
- ERβ:
-
Estrogen receptor beta
- MOI:
-
Multiplicity of infection
- mTOR:
-
Mammalian target of rapamycin
- PI3K:
-
Phosphoinositide-3-kinase
- p-mTOR:
-
phosphorylated-mTOR
- RT-qPCR:
-
Reverse transcription quantitative polymerase chain reaction
- SD:
-
Standard deviation
References
Moore DD, Luu HH. Osteosarcoma. Cancer Treatment Res. 2014;162:65.
Ottaviani G, Jaffe N. The Epidemiology of Osteosarcoma. In: Jaffe N., Bruland O., Bielack S. (eds) Pediatric and Adolescent Osteosarcoma. Cancer Treatment and Research, 2009, vol 152. Springer, Boston, MA. https://xs.scihub.ltd/https://doi.org/10.1007/978-1-4419-0284-9_1.
Isakoff MS, et al. Osteosarcoma: current treatment and a collaborative pathway to success. Journal of clinical oncology. 2015;33(27):3029.
Anderson ME. Update on survival in osteosarcoma. Orthopedic Clinics. 2016;47(1):283–92.
Zhang J, et al. MicroRNAs in osteosarcoma. Clinica Chimica Acta. 2015;444:9–17.
Zheng K, et al. Dual actions of albumin packaging and tumor targeting enhance the antitumor efficacy and reduce the cardiotoxicity of doxorubicin in vivo. Int J Nanomed. 2015;10(10):5327.
Younes M, Honma N. Estrogen receptor β. Arch Pathol Lab Med. 2011;135(1):63–6.
Deroo BJ, Korach KS. Estrogen receptors and human disease. J Clin Investigation. 2006;116(3):561–70.
Shughrue PJ, Lane MV, Merchenthaler I. Comparative distribution of estrogen receptor-α and-β mRNA in the rat central nervous system. J Comparative Neurol. 1997;388(4):507–25.
Pierdominici M, et al. Estrogen receptor β ligation inhibits Hodgkin lymphoma growth by inducing autophagy. Oncotarget. 2017;8(5):8522.
Yang M, et al. Estrogen receptor β exhibited anti-tumor effects on osteosarcoma cells by regulating integrin, IAP, NF-kB/BCL-2 and PI3K/Akt signal pathway. J Bone Oncol. 2017;9:15–20.
Motylewska E, Stasikowska O, Mełeń-Mucha G. The inhibitory effect of diarylpropionitrile, a selective agonist of estrogen receptor beta, on the growth of MC38 colon cancer line. Cancer Letters. 2009;276(1):68–73.
Jacome LF, et al. Estradiol and ERβ agonists enhance recognition memory, and DPN, an ERβ agonist, alters brain monoamines. Neurobiol Learning Memory. 2010;94(4):488–98.
CARO LHP, et al. 3-Methyladenine, an inhibitor of autophagy, has multiple effects on metabolism. Eur J Biochem. 1988;175(2):325–9.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4):402–8.
Dey P, et al. Insight into the mechanisms of action of estrogen receptor β in the breast, prostate, colon, and CNS. J Mol Endocrinol. 2013;51(3):T61–74.
Ma R, et al. Estrogen Receptor β as a Therapeutic Target in Breast Cancer Stem Cells. J Natl Cancer Institute. 2017;109(3):djw236. https://doi.org/10.1093/jnci/djw236.
Chaurasiya S, et al., Estrogen receptor β exerts tumor suppressive effects in prostate cancer through repression of androgen receptor activity. bioRxiv, 2019: p. 850180. https://www.biorxiv.org/content/10.1101/850180v1.full.
Liu J, et al. Therapeutic utility of natural estrogen receptor beta agonists on ovarian cancer. Oncotarget. 2017;8(30):50002.
Arabacı Tamer S, et al. Treatment with estrogen receptor agonist ERβ improves torsion-induced oxidative testis injury in rats. Life Sciences. 2019;222:203–11.
Dupuis ML, et al. The Natural Agonist of Estrogen Receptor β Silibinin Plays an Immunosuppressive Role Representing a Potential Therapeutic Tool in Rheumatoid Arthritis. Front Immunol. 2018;9:1903. https://doi.org/10.3389/fimmu.2018.01903.
Wei Y, et al. Estrogen Receptor Beta (ERβ) Mediated-CyclinD1 Degradation via Autophagy Plays an Anti-Proliferation Role in Colon Cells. Int J Biol Sci. 2019;15(5):942–52.
Wei Y, et al. ERβ promotes Aβ degradation via the modulation of autophagy. Cell Death Disease. 2019;10(8):565.
Doherty J, Baehrecke EH. Life, death and autophagy. Nature Cell Biol. 2018;20(10):1110–7.
Dikic I, Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol. 2018;19(6):349–64.
Huang R, et al. Deacetylation of Nuclear LC3 Drives Autophagy Initiation under Starvation. Molecular Cell. 2015;57(3):456–66.
Jiang P, Mizushima N. LC3- and p62-based biochemical methods for the analysis of autophagy progression in mammalian cells. Methods. 2015;75:13–8.
Moscat J, Karin M, Diaz-Meco MT. p62 in Cancer: Signaling Adaptor Beyond Autophagy. Cell. 2016;167(3):606–9.
Xu XW, et al. SP1 reduces autophagic flux through activating p62 in gastric cancer cells. Mol Med Reports. 2018;17(3):4633–8.
Zhang H, et al. DEAD Box Protein 5 Inhibits Liver Tumorigenesis by Stimulating Autophagy via Interaction with p62/SQSTM1. Hepatology. 2019;69(3):1046–63.
Paquette M, El-Houjeiri L, Pause A. mTOR Pathways in Cancer and Autophagy. Cancers. 2018;10(1):18.
Hu K, Dai H-B, Qiu Z-L. mTOR signaling in osteosarcoma: Oncogenesis and therapeutic aspects. Oncology reports. 2016;36(3):1219–25.
Kim S-H, et al. Deoxypodophyllotoxin induces cytoprotective autophagy against apoptosis via inhibition of PI3K/AKT/mTOR pathway in osteosarcoma U2OS cells. Pharmacol Reports. 2017;69(5):878–84.
Li Z, et al. Honokiol induces autophagy and apoptosis of osteosarcoma through PI3K/Akt/mTOR signaling pathway. Mol Med Reports. 2018;17(2):2719–23.
Acknowledgements
None.
Funding
None.
Author information
Authors and Affiliations
Contributions
Conception and design of the research: HT; acquisition of data: MY; analysis and interpretation of data: BL; statistical analysis: WY; drafting the manuscript: ZY; revision of the manuscript for important intellectual content: ZY. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Yang, Z., Yu, W., Liu, B. et al. Estrogen receptor β induces autophagy of osteosarcoma through the mTOR signaling pathway. J Orthop Surg Res 15, 50 (2020). https://doi.org/10.1186/s13018-020-1575-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13018-020-1575-1