Abstract
Background
Carbapenemase genes are one of the most frequent mechanisms reported in carbapenem-resistant P. aeruginosa; however, description of P. aeruginosa co-harbouring two or more carbapenemases is unusual.
Methods
In this study we evaluated the presence of carbapenemase genes and the clonality of P. aeruginosa isolates obtained from a hospital over a 12-year period. A total of 127 isolates of carbapenem-resistant P. aeruginosa recovered from 109 patients feces (four samples), rectal swab (three samples), nasal swab (one sample) and anal abscess (one sample), were evaluated. Minimum inhibitory concentrations of the following antibiotics imipenem, meropenem and polymyxin E were determined by broth microdilution. The molecular profile of isolates was evaluated by pulsed field gel electrophoresis (PFGE). PCR for the following carbapenemase genes blaIMP;blaSPM;blaVIM;blaSIM;blaNDM;blaKPC;blaGES and nucleotide sequencing to confirm the enzyme gene types were performed and compared with the database available on the Internet (BLAST-http://www.ncbi.nlm.nhi.gov/blast/).
Results
All isolates were carbapenem-resistant, their MIC50 and MIC90 were respectively 64 μg/mL and 256 μg/mL to imipenem and 32 μg/mL and 256 μg/mL to meropenem, all isolates except one (MIC = 8 mg/L) were susceptible to polymyxin E. The most frequent carbapenemase genes identified were blaSPM identified in 41 isolates (32%), followed by 10 with blakpc and 5 with blaVIM (3.9%). All belonged to the class SPM-1 and VIM-2. In 2011, one isolate harbouring three carbapenemase genes (SPM-1, VIM-2 and KPC-2) that belonged to a new clone was identified in a hematopoietic stem cell transplanted patient. Then, 19 carbapenem-resistant P. aeruginosa were identified in an outbreak that occurred in the bone marrow transplant unit, all positive for SPM-1 gene, and 9 (47.3%) harbored both SPM-1 and KPC.
Conclusion
Our findings showed that PCR for KPC gene should be performed to evaluate carbapenem resistance in P. aeruginosa and that this agent can harbor more than one carbapenemase gene. Attention should be focused on the possible rapid spread of KPC in P. aeruginosa isolates and for the fact that P. aeruginosa may become a reservoir of this transmissible resistance mechanism.
Similar content being viewed by others
Background
Carbapenem resistant P. aeruginosa has become an important problem all over the world challenging the current diagnostic approaches. Carbapenemase genes are one of the most frequent mechanisms reported in carbapenem-resistant P. aeruginosa[1]-[5]. It is important to identify carbapenemase genes transmitted on mobile genetic elements which can lead to the spread of resistance of P. aeruginosa to carbapenem, which are the main drugs used to treat infections caused by this agent. In Brazil, the most common carbapenemase is the metallo-betalactamase, SPM, however, recently P. aeruginosa harboring KPC was identified [2].
In this study, we evaluated the presence of carbapenemase genes and the clonality of carbapenem- resistant P. aeruginosa isolates obtained from a teaching hospital over a 12-year period.
Methods
Study setting
The study was conducted in The Central Institute of Hospital das Clinicas (ICHC FMUSP), Brazil, a teaching hospital with 1,000 beds, ten intensive care units totalizing 110 beds and a bone marrow transplant ward with 20 beds.
Isolates
A total of 129 P. aeruginosa carbapenem-resistant clinical isolates identified over a 12-year period, from 1998 to 2012, recovered from 109 patients, hospitalized in the Clinical and Surgical nursery, Intensive Care, Burned, Haematology and Bone Marrow units at Hospital das Clinicas-FMUSP were evaluated.
Susceptible profile
Minimum inhibitory concentrations (MICs) of imipenem, meropenem and polymixin E were determined by broth microdilution according with Clinical Laboratory Standards Institute (CLSI 2012).
Molecular typing
Bacterial isolates were grown on blood agar overnight at 37°C. Gel blocks were made by using equal volumes of 2% low-melting-point agar (BioRad, USA) and a bacterial suspension of 9*108 cells. Genomic DNA was digested with 10U of XBAI (Fermentas, USA), [6] (Sekiguchi ref). DNAs were separated by pulsed-field gel electrophoresis (PFGE) using a CHEF-DR III system (Bio-Rad, USA). Running conditions were 21h at 14°C, with and initial switching time of 1 s and final time of 30 s, at 6V/cm. PFGE patterns were interpreted according to Tenover et al. 1997 [7].
Carbapenemases genes
PCR for the following carbapenemase genes blaIMP;blaSPM;blaVIM;blaSIM;blaNDM;blaKPC;blaGES was done as described previously [8]-[11]. The following reference strains were used as control in this study: P. aeruginosa that produced IMP-1, VIM-2, SIM-1, SPM-1 [10], KPC ATCC and E. coli NDM. Nucleotide sequencing to confirm the enzyme gene types was performed by MegaBACE 1000. The sequences were analyzed using the software Sequence Analyzer with the Base Caller Cimarron 3.12. The genetic sequence was compared with the database available on the Internet (BLAST-http://www.ncbi.nlm.nhi.gov/blast/). The KPC sequence was also comparing with KPC lahey databases (http://www.lahey.org/Studies/other.asp#table1).
Results
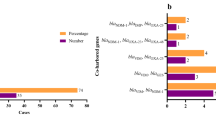
All isolates were carbapenem-resistant, their MIC50 and MIC90 were respectively 64μg/mL and 256μg/mL to imipenem and 32μg/mL and 256μg/mL to meropenem. All isolates except one (MIC: 8μg/mL), were susceptible to colistin, MICs varied from 0.25 to 2μg/mL. They were recovered from blood (120 samples), feces (four samples), rectal swab (three samples), nasal swab (one sample) and anal abscess (one sample). PFGE showed that, from 1998 to 2009, 25% (27 of 108 strains) belonged to one predominant clone (A1), thirty-two (A2) of 108 isolates (29.7%) were closely related to them and sixteen (14.8%) were possibly related (A3, A4, A5, A6 and A7) to the predominant clone (A1). Thirty-three (30.5%) showed no relation with the predominant clone A1. Among the 129 isolates 50 (39%) harboured a carbapenemase, the most frequent carbapenemase genes identified were blaSPM identified in 41 isolates (32%) (Figure11), followed by 10 with blakpc (Figure22) and 5 with blaVIM (Figure33). GES-5 was identified only in 3 isolates from one burned patient. Pseudomonas aeruginosa harbouring SPM-1 was identified for the first time in the Bone Marrow Transplant unit in 1998, VIM-2 in the Emergency Room in 2001 and GES-5 in the Burned Intensive Care Unit. GenBank accession numbers: JX840596 (VIM-2), JX682700-JX682705 (KPC-2) and JX870518-JX870528 (SPM-1).
In 2012 during an outbreak that occurred in the bone marrow transplant unit, 19 carbapemem-resistant P. aeruginosa were identified (clone K, L, L1, L2, L3 and M), all positive for SPM-1 gene, and 10 (47.3%) harboured both SPM-1 and KPC-2. The majority of the KPC, six isolates of nine, belong to the clone of the outbreak (K).
The relation between molecular profile and carbapenemase gene for all period is shown on Table 11.
Discussion
It is the first report of P. aeruginosa co-harbouring blaKPC and blaSPM genes and the first report in Brazil of P. aeruginosa carrying KPC-2, VIM-2 and SPM-1. SPM-1 was the most frequent carbapenemase identified in our hospital, followed by KPC-2. KPC in P. aeruginosa is rare and occurs mainly in the American continent [1]-[5],[12]. Recently, P. aeruginosa harbouring KPC was described in Argentina [13] and Iran [14], showing the potential rapid dissemination of this mechanism of resistance to the world.
Even though P. aeruginosa harbouring KPC was identified in 2010 in Brazil [2], no other report was published since then.
Only 50 of 129 P. aeruginosa carbapenem-resistant harboured a carbapenemase evaluated in this study. Thus, the carbapenem resistance could be related to other mechanism of resistance such as outer-membrane protein alteration, efflux system overexpression or new carbapenemase not yet identified [15]-[17].
Pseudomonas aeruginosa co-harbouring carbapenemase is uncommon; there are few reports in the literature [1;3]. Pseudomonas aeruginosa isolate co-harbouring KPC and a metallo-β-lactamase (IMP-8) was recently reported in Puerto Rico [3] and isolates co-harbouring KPC and VIM gene were identified in Colombia [1]. We described a new clone of P. aeruginosa co-harbouring SPM-1 and KPC-2 that caused an outbreak in a Bone Marrow transplantation unit, and an isolate co-harbouring three carbapenemase (SPM-1; KPC-2 and VIM-2) that belonged to a different clone then previous described in two outbreaks that occurred in this unit and were controlled with reinforcement of hand hygiene and contact precautions, one due to P. aeruginosa harbouring SPM-1 and other harbouring VIM-2 [18].
This P. aeruginosa isolate harbouring KPC-2 was identified for the first time in our hospital in 2011 in a hematopoietic stem cell transplanted patient. It harboured three carbapanemase genes (SPM-1, VIM-2 and KPC-2), and belonged to a new clone (JA) not identified before in the hospital. This isolate showed a resistant profile to both imipenem and meropenem with MIC of 64μg/mL and 32μg /mL, respectively, but was susceptible to polymyxin and colistin with a MIC of 2μg /mL for both drugs.
In 2012 during an outbreak that occurred in the Bone Marrow Transplant unit, 19 carbapemem-resistant P. aeruginosa were identified (clone K, L, L1, L2, L3 and M), all positive for SPM-1 gene, and 10 (47.3%) harbored both SPM-1 and KPC-2. The majority of the KPC, six isolates of ten, belong to the clone of the outbreak (KB). Our data showed different clones circulating in our hospital and a new one predominate clone harboring KPC. A recent study also described dissemination of a new clone of P. aeruginosa harbouring KPC in a hospital in Argentina after a K. pneumonia- KPC positive outbreak [13].
Other interesting findings of our study are that GES-5 was restricted to the Burned Intensive Care Unit and was identified only in 2001. VIM-1 was first identified in the Emergency Room and then in the Bone Marrow Transplant Unit, and SPM is spread in different units in the hospital. These results were similar with previous Brazilian reports that showed that SPM-1 is endemic in several hospitals in the country [19],[20].
The complete sequence of two KPC-harbouring plasmids, Plasmid pCOL-1 (31529bp), IncP-6 replicon group and Plasmid pPA-2 (7995bp) from P. aeruginosa showed that they differing in size and in incompatibility group, and harbouring different genetic structures containing the blaKPC-2 genes [21]. These findings suggest that the carbapenemase resistance dissemination due to KPC in P. aeruginosa will be similar to that seen in Enterobacteriaceae. Thus, it is very important to understand the epidemiology of these multiresistant isolates, in order to achieve early implementation of adequate control measures to contain and reduce their dissemination in the hospital setting. Pseudomonas aeruginosa can acquired this transmissible resistance mechanism, going unnoticed and be a source of spread of KPC to other genus and species of bacteria. Besides carbapenem-resistance in P. aeruginosa can be due to two or more carbapenemase genes, including KPC-gene.
In conclusion, our findings showed that SPM-1 is the most frequent carbapenemase identified in our hospital, followed by KPC-2. Thus, PCR for KPC gene should be performed to evaluate carbapenem resistance in P. aeruginosa and this agent can harbor more than one carbapenemase gene. Attention should be focused on the possible rapid spread of KPC in P. aeruginosa isolates and for the fact that P. aeruginosa may become a reservoir of this transmissible resistance mechanism.
References
Correa A, Montealegre MC, Mojica MF, Maya JJ, Rojas LJ, De La Cadena EP, Ruiz SJ, Recalde M, Rosso F, Quinn JP, Villegas MV: First report of a pseudomonas aeruginosa isolate co-harboring KPC and VIM carbapenemases. Antimicrob Agents Chemother 2012,56(10):5422-5423. 10.1128/AAC.00695-12
Jcome PR, Alves LR, Cabral AB, Lopes AC, Maciel MA: First report of KPC-producing pseudomonas aeruginosa in Brazil. Antimicrob Agents Chemother 2012, 56(9):4990.
Martnez T, Vzquez GJ, Aquino EE, Ramrez-Ronda R, Robledo IE: First report of a Pseudomonas aeruginosa clinical isolate co-harboring KPC-2 and IMP-18 carbapenemases. Int J Antimicrob Agents 2012,39(6):542-543. 10.1016/j.ijantimicag.2012.02.009
Cuzon G, Naas T, Villegas MV, Correa A, Quinn JP, Nordmann P: Wide dissemination of Pseudomonas aeruginosa producing beta-lactamase blaKPC-2 gene in Colombia. Antimicrob Agents Chemother 2011,55(11):5350-5353. 10.1128/AAC.00297-11
Villegas MV, Lolans K, Correa A, Kattan JN, Lopez JA, Quinn JP: First identification of Pseudomonas aeruginosa isolates producing a KPC-type carbapenem-hydrolyzing beta-lactamase. Antimicrob Agents Chemother 2007,51(4):1553-1555. 10.1128/AAC.01405-06
Sekiguchi J, Asagi T, Miyoshi-Akiyama T, Fujino T, Kobayashi I, Morita K, Kikuchi Y, Kuratsuji T, Kirikae T: Multidrug-resistant Pseudomonas aeruginosa strain that caused an outbreak in a neurosurgery ward and its aac(6')-Iae gene cassette encoding a novel aminoglycoside acetyltransferase. Antimicrob Agents Chemother 2005,49(9):3734-3742. 10.1128/AAC.49.9.3734-3742.2005
Tenover F, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B: Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strains typing. J Clin Microbiol 1995,33(9):2233-2239.
Bradford PA, Bratu C, Urban C, Visalli M, Mariano N, Landman D, Rahal JJ, Brooks S, Cebular S, Quale J: Emergence of carbapenem-resistant Klebsiella species possessing the class A carbanem hydrolyzing KPC-2 and Inhibithor-resitant TEM30 β-lactamases in New York. Clin Infect Dis 2004,39(1):55-60. 10.1086/421495
Chen Y, Zhou Z, Zang Y, Yu Y: Emergence of NDM-1 producing Acinetobacter baumannii in China. J Antimicrob Chemother 2011,66(6):1255-1259. 10.1093/jac/dkr082
Mendes ER, Kiyota KA, Monteiro J, Castanheira M, Andrade SS, Gales AC, Pignatari AC, Tufik S: Rapid detection and identification of Metallo-β-lactamase- encoding genes by multiplex real time PCR assay and melt curve analysis. J Clin Microbiol 2007,45(2):544-547. 10.1128/JCM.01728-06
Kim SY, Park YJ, Yu JK, Kim HS, Park YS, Yoon JB, Yoo JY, Lee K: Prevalence and mechanisms of decreased susceptibility to carbapenems in Klebsiella pneumoniae isolates. Diagn Microbiol Infect Dis 2007,57(1):85-91. 10.1016/j.diagmicrobio.2006.05.008
Garca Ramrez D, Nicola F, Zarate S, Relloso S, Smayevsky J, Arduino S: Emergence of Pseudomonas aeruginosa with KPC-type carbapenemase in a teaching hospital: an 8-year study. J Med Microbiol 2013, 62: 1565-1570. 10.1099/jmm.0.059923-0
Santella G, Cittadini R, Papalia M, Vera Ocampo C, Del Castillo M, Vay C, Gutkind G, Radice M: First clonal spread of KPC-producing Pseudomonas aeruginosa in Buenos Aires, Argentina. Infect Genet Evol 2012,12(8):2003-2005. 10.1016/j.meegid.2012.03.022
Lari AR, Azimi L, Rahbar M, Alaghehbandan R, Sattarzadeh-Tabrizi M: First report of Klebsiella pneumonia carbapenemase-producing Pseudomonas aeruginosa isolated from burn patients in Iran: phenotypic and genotypic methods. GMS Hyg Infect Control 2014, 9(1):Doc06.
Liu Y, Li XY, Wan LG, Jiang WY, Li FQ, Yang JH: Efflux system overexpression and decreased OprD contribute to the carbapenem resistance among extended-spectrum beta-lactamase-producing Pseudomonas aeruginosa isolates from a Chinese university hospital. Microb Drug Resist 2013,19(6):463-468. 10.1089/mdr.2013.0010
Castanheira M, Deshpande LM, Costello A, Davies TA, Jones RN: Epidemiology and carbapenem resistance mechanisms of carbapenem-non-susceptible Pseudomonasaeruginosa collected during 2009-11 in 14 European and Mediterranean countries. J Antimicrob Chemother 2014,69(7):1804-1814. 10.1093/jac/dku048
Aghazadeh M, Hojabri Z, Mahdian R, Nahaei MR, Rahmati M, Hojabri T, Pirzadeh T, Pajand O: Role of efflux pumps: MexAB-OprM and MexXY(-OprA), AmpC cephalosporinase and OprD porin in non-metallo-β-lactamase producing Pseudomonas aeruginosa isolated from cystic fibrosis and burn patients. Infect Genet Evol 2014, 24: 187-192. 10.1016/j.meegid.2014.03.018
Paez J, Levin AS, Fu L, Basso M, Fonseca GH, Dulley FL, Rossi F, Guimares T, Costa SF: Clusters of infection due to metallo-β-lactamase-producing Pseudomonas aeruginosa in stem cell transplant and haematology units. J Hosp Infect 2011,77(1):76-77. 10.1016/j.jhin.2010.08.007
Camargo CH, Bruder-Nascimento A, Mondelli AL, Montelli AC, Sadatsune T: Detection of SPM and IMP metallo-β-lactamases in clinical specimens of Pseudomonas aeruginosa from a Brazilian public tertiary hospital. Braz J Infect Dis 2011,15(5):478-481. 10.1016/S1413-8670(11)70231-8
Scheffer MC, Gales AC, Barth AL, Carmo Filho JR, Dalla-Costa LM: Carbapenem-resistant Pseudomonas aeruginosa: clonal spread in southern Brazil and in the state of Gois. Braz J Infect Dis 2010,14(5):508-509.
Naas T, Bonnin RA, Cuzon G, Villegas MV, Nordmann P: Complete sequence of two KPC-harbouring plasmids from Pseudomonas aeruginosa. J Antimicrob Chemother 2013,68(8):1757-1762. 10.1093/jac/dkt094
Funding section
This study was performed using internal funding.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Concept and design (SFC), data collection and laboratory work (CR, LF, JR, GL, LCS) data analysis and interpretation (CR, ASL, SFC), drafting of the manuscript (CR,TG, ASL, SFC), critical review of the manuscript (CR, TG, SL, SFc), final approval of manuscript for publication (CR, LF, JR, GL, LCS,TG, ASl, SFC). All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Rizek, C., Fu, L., dos Santos, L.C. et al. Characterization of carbapenem-resistant Pseudomonas aeruginosa clinical isolates, carrying multiple genes coding for this antibiotic resistance. Ann Clin Microbiol Antimicrob 13, 43 (2014). https://doi.org/10.1186/s12941-014-0043-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12941-014-0043-3