Abstract
Background
In light of rampant childhood diarrhoea, this study investigated bacterial pathogens from human and non-human sources in an urban informal settlement.
Meat from informal abattoirs (n = 85), river water (n = 64), and diarrheic stool (n = 66) were collected between September 2015 and May 2016. A duplex real-time PCR, gel-based PCR, and CHROMagar™STEC were used to screen Tryptic Soy Broth (TSB) for diarrheic E. coli. Standard methods were used to screen for other selected food and waterborne bacterial pathogens.
Results
Pathogens isolated from stool, meat, and surface water included Salmonella enterica (6, 5, 0%), Plesiomonas shigelloides (9, 0, 17%), Aeromonas sobria (3, 3, 0%), Campylobacter jejuni (5, 5, 0%), Shigella flexneri (17, 5, 0%), Vibrio vulnificus (0, 0, 9%), and diarrheic E. coli (21, 3, 7%) respectively. All the isolates were resistant to trimethoprim–sulphamethoxazole.
Conclusions
There was a high burden of drug resistant diarrheal pathogens in the stool, surface water and meat from informal slaughter. Integrated control measures are needed to ensure food safety and to prevent the spread of drug resistant pathogens in similar settings.
Similar content being viewed by others
Background
Diarrheal disease is a major cause of morbidity in the Western Cape province and the city of Cape Town in South Africa [1]. Infant mortality from diarrhea in South Africa is characterized by a seasonal unimodal peak from March – June each year [2]. In the Western Cape and Cape Town, the peak is experienced around March [3]. A recent South African Demographic and Health Survey (2016) showed that 10% of children under five years had experienced diarrhea in the preceding two weeks; 63% of these sought medical treatment [4]. Diarrhea is the second leading cause of under-five mortality in South Africa (16%), trailing behind HIV/AIDS (20.1%) [5]. In the Western Cape Province, diarrhea is the 3rd leading cause of under-five mortality (11%); almost half (42.9%) of child diarrheal deaths in the Cape Town metro sub-district occur at home [3]. Among those that seek primary healthcare, some are locally managed, while others are referred to tertiary care; a main contributing factor for diarrheal death among those referred to tertiary care in South Africa is a failure to correctly assess the severity of the child’s condition at the primary care level [6]. Of those hospitalized, the under 5 case fatality rate attributable to diarrhea is estimated at 7.3% [6].
Public primary health care practice in South Africa relies on the syndromic management of diarrheal disease [7]. The South African Standard Treatment Guidelines (STGs) for acute diarrhoea in children recommend rehydration and a single dose of intramuscular ceftriaxone at 80 mg/kg for infants (< 4 weeks old), the malnourished, and where danger signs exist. Explicitly, for dysentery, the STGs indicate treatment with oral ciprofloxacin (15 mg/kg 12 hourly for three days) followed by treatment for amoebic dysentery if symptoms do not improve after three days [8].
Supranational surveillance projects such as the Global Enteric Multicenter Study (GEMS) [9] have provided relevant information on the common causes of paediatric infectious diarrhea in sub-Saharan Africa but demonstrated that public health interventions should be based on locally generated data [10]. A better understanding of the local environmental factors that favour transmission of enteric pathogens is particularly crucial for vulnerable communities such as those living in poverty in low-cost housing settlements [11]. Given the unhygienic conditions at local abattoirs, improper disposal of abattoir and domestic waste, and the reports of childhood diarrhea, we set out to identify the enteric bacterial pathogens present in an urban informal settlement in Cape Town. We therefore screened children that presented with diarrhea at the local Community Health Center, water from the local canal, and meat from the local informal food trade.
Methods
Setting
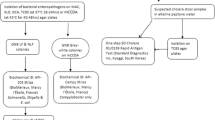
The City of Cape Town is temperate with moderately wet winters and dry, warm summers. The Nyanga urban informal settlement surveyed is comprised of ~ 16,000 dwellings with ~ 58,000 residents. The population density is estimated at 19,000/km2. Eighty-one percent of households have access to communal flush toilets connected to a sewer system, while 11% rely on bucket toilets; and 3% report no access to toilet facilities. Only 53.5% of households have piped water inside their dwellings [12]. The Nyanga settlement is located in a seasonal wetland that has been canalized subsequent to urbanization resulting in a canalized river flowing through this densely populated community (Fig. 1). Informal abattoirs are an important component of the micro-economy and include the street-side slaughter of livestock and the selling of meat and meat products.
Patient selection and specimen sampling
The caregivers of children (under 12 years of age) with diarrhoea, attending the local Community Health Center (CDC) on Monday, Wednesday, and Friday of every week between October 2015 and April 2016, were approached at random for enrolment after informed consent was obtained. Clinical and epidemiological data was collected from consenting caregivers. Freshly passed stool was collected using a screw-capped stool collection container (Sarstedt, Nümbrecht, Germany) from the diaper (or a triple tissue layer). Meat samples (raw and roasted chicken, beef, and pork) from all road-side stalls in the community were collected randomly in sterile stomacher bags (Tekmar, Cincinnati, OH) on the first two Fridays of every month using the guidelines for integrated surveillance of foodborne antimicrobial resistance [13]. Surface water samples were collected from four points (LR13, LR14, LR15, and LR16) once every two weeks for eight months along the canalized river using the depth-integrated grab sampling method at a depth of 30 cm below the surface towards the center of the stream [14]. Samples were transported to the laboratory within 12 h of the collection in a 4 °C temperature – monitored box.
This non-interventional survey study was approved by the Human Subjects Research Ethics Committee of the University of Cape Town (HREC 2015/140) and the City of Cape Town Health Department, Klipfontein Health sub-district (6547–10,516).
Informed consent: “Informed consent was obtained from all individual participants included in this study.
Sample processing
A pea-sized amount of stool, 25 g of meat tissue (pummeled using a sterile mortar and pestle), and the filtrate (on a 0.45 μm membrane filter) of 100 ml of surface water were inoculated in 90 ml, 225 ml, and 90 ml of TSB (Oxoid, Basingstoke, UK) respectively and incubated at 35 °C overnight as previously described [15, 16]. From overnight enrichments, a loopful was streaked on CHROMagar™STEC (CHROMagar Microbiology, Paris, France) for STEC, Xylose Lysine Deoxycholate (XLD, Greenpoint Media Laboratory, NHLS, Albertynshof) for Shigella and Salmonella, and Cefsulodin -Irgasan-Novobiocin (CIN, Greenpoint Media Laboratory, NHLS, Albertynshof) for Aeromonas and Yersinia with overnight incubation at 35 °C. For Campylobacter, samples (1 ml of surface water, 1 ml of tissue rinsate in TSB, or a loopful of stool sample) were directly streaked onto Charcoal Cefoperazone Deoxycholate modified Agar (CCDA) and incubated under microaerophilic conditions for 72 h at 35 °C. To culture Vibrio, 10 ml of water or 1-10 ml of stool was inoculated in 90 ml of Alkaline Peptone Water and incubated at 42 °C for eight hours. Subsequently, a loopful of the surface pellicle was sub-cultured onto a Thiosulfate – Citrate – Bile salts – Sucrose (TCBS) agar and incubated overnight at 35 °C. For P. shigelloides, a loopful of TSB was used to inoculate MacConkey agar with crystal violet (McA) and Cefsulodin – Irgasan - Novobiocin (CIN) agar and incubated at 35 °C for 24 h. Non- lactose fermenting colonies on McA and opaque colonies with a pink center on CIN, were then tested for oxidase activity before confirmatory identification. A maximum of five mauve colonies from each CHROMagar™STEC plate were sub-cultured separately on McA agar with crystal violet and sorbitol MacConkey agar plates. Suspect colonies from the different selective media were picked for confirmatory identification and characterisation. All isolates were identified, and their minimum inhibitory concentration (MIC) values determined using VITEK 2 (bioMérieux, USA). E. coli isolates from CHROMagar™STEC were tested for the presence of stx using real-time PCR on a LightCycler®480 II Instrument (Roche Life Sciences, Industriestrasse, Switzerland) using LightCycler® 480 Probes Master mastermix and primers as previously described. (Table 1) [18].
The presence of the fimbrial adhesion gene (daaC) for diffusely adherent E. coli (DAEC), the anti-aggregation protein transporter gene (aat) for enteroaggregative E. coli (EAggEC) heat-stable (ST) and heat-labile (LT) enterotoxin genes for enterotoxigenic E. coli (ETEC), the invasive plasmid antigen (ipa) gene for enteroinvasive E. coli (EIEC), the bundle-forming pili (bfp) gene for typical enteropathogenic E. coli (EPEC) and the intimin coding gene (eae) for EPEC were determined using end-point PCR on an Applied Biosystems™ 2720 Thermal Cycler platform using the QIAGEN PCR-multiplex kit (QIAGEN GmbH, Hilden, Germany) followed by agarose gel detection as previously described [19], using primers as shown in Table 2 (Inqaba Biotec Laboratory, South Africa).
The positive control strains that were used for the different diarrheic E. coli pathotypes are shown below (Table 3) ([23]):
Similarly, sample TSB enrichment broths from all sources were subjected to nucleic acid extraction using the MagNApure bacterial/fungal DNA extraction kit on a MagNApure LC instrument (Roche Diagnostics, Industriestrasse, Switzerland). PCR testing was done as described above [24].
Shigella were serotyped using the Wellcolex* colour Shigella Rapid latex agglutination test (Oxoid, Basingstoke, UK), while Salmonella and E. coli (O-typing) [25] were serotyped at the National Institute for Communicable Disease (NICD), Sandringham, Johannesburg using standard serological methods.
Enumeration of faecal coliforms in water
Six ten-fold serial dilutions of 10 ml of canalized water in 90 ml of sterile water were filtered through 0.45 μM nitrocellulose membranes (Pall Corporation, Port Washington, USA) under vacuum suction and incubated on modified mTEC agar (Difco, Detroit, USA) at 35 °C for two hours and later at 44 °C for 22 h as previously described, followed by manual counting of colonies [16].
Data analysis
Data on clinical and epidemiological characteristics were analyzed using Epi Info 7™ (CDC, Atlanta, USA). Comparison of proportions was done using a two-tailed Chi-square test or Fischer’s exact test, with p < 0.05 considered significant. MIC data were analyzed using the WHONET version 5.6 software (WHO, Geneva, Switzerland).
Results
Stool specimens were collected from 66 children with diarrhoea (mean age ~ 15 months, range 2–36 months), 85 meat samples and 64 community canal water samples. Thirty-nine percent of children had received rotavirus immunisation. This study targeted patients three years and younger that presented with diarrhoea to the local centre during the study period. Among the children that presented with diarrhoea, 5% (3/66), 20% (13/66), and 17% (11/33) reported bloody stool, vomiting, and fever respectively (Table 4).
Overall, an enteric bacterial pathogen was isolated from 55% (36/66) of the cases; in 17% (6/36) of these, more than one bacterial pathogen was isolated (Shigella flexneri and STEC-one patient, Shigella flexneri and Campylobacter jejuni-one patient, Aeromonas sobria and DAEC-one patient, Shigella flexneri and DAEC-one patient, DAEC and EAEC-one patient, DAEC and EAggEC-one patient). There was a high prevalence of S. flexneri (Table 5) and DAEC (Table 6) in stool. Blood in stool was reported in cases where STEC (1/1), A. sobria (1/2), and DEC (1/14) were isolated. Notably, none (0/11) of the S. flexneri cases presented with bloody stool.
The various raw meat samples (chicken, beef, pork, and mutton) that were tested carried various bacterial pathogens which included diarrheic E. coli, C. jejuni, S. enterica serotype Idikan, A. sobria and P. shigelloides. None of the targeted bacterial pathogens were isolated from any of the roasted meat samples. No tellurite resistant STEC, EAggEC, ETEC, DAEC or EIEC were isolated from any of the meat samples in this setting (Table 6).
For all the pathotypes, more of the virulence marker genes were detected (Fig. 2) in TSB enrichment of stool as compared to the actual number of diarrheic E. coli pathotypes isolated on CHROMagar™STEC. This could be explained by the fact that not all diarrheic E. coli are tellurite resistant and so would not form colonies on CHROMagar™STEC.
Electrophoresis gel image for PCR product following multiplex PCR ampification for aat, ipa, and daaC. Lanes 1 and 17 contained the 1 kb ladder (GeneRuler™ DNA molecular weight ladder -ThermoFisher Scientific Inc., MA, USA). Lanes 2–32 contained samples except for lanes 14 (positive control for aat),18 (positive control for ipa), 19 (negative control), 32 (positive control for daaC). Samples in lanes 9, 10, 13, 21, 22, 23, 24, 25, 26, and 29 were positive for daaC. The sample in lane 30 was positive for ipa. The rest of the samples were negative for aat, ipa, and daaC. The gel image was captured using the E-Gel GelCapture Software (version 1.7)
Nevertheless, there was a high prevalence of stx1 (20%, 17/85) in TSB enrichment broths of these samples. For raw beef, S. enterica, P. shigelloides, A. sobria, and EPEC were isolated. For raw pork, only DAEC was isolated while S. enterica and C. jejuni were isolated from raw chicken (Table 7).
Surface water from the community canal (at sampling points LR13, LR14, LR15, and LR16) yielded high numbers of faecal coliforms (mean CFU/ml =2.74E+ 05–1.11E+ 06) with the highest levels seen in February (p = < 0.001), which was also the driest (zero precipitation) and hottest month; precipitation in March 2016 coincided with the lowest measured surface water contamination level (Fig. 3). P. shigelloides (11/64, 17%) was the most abundant enteric bacterial pathogen in water; others included S. flexneri, V. vulnificus, A. sobria, EAggEC, STEC, and DAEC. The samples, however, had a high prevalence of daaC (38%, 24/64) and stx1 (30%, 19/64).
Mean faecal coliform counts (CFU/ml) for four water collection points along the Lotus River (LR13, LR14, LR 15, and LR 16), monthly average rainfall (mm), and average daily minimum temperatures (°C) between July 2015 and April 2016. The Figure was developed using the Microsoft Office Excel 365 ProPlus, version 1902
For all sample types, diarrheic E. coli virulence genes were detected more frequently with direct testing from TSB, compared to isolation from CHROMagar™STEC. Of note, all isolates on CHROMagar™STEC yielded corresponding PCR targets on direct TSB PCR.
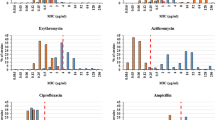
Antimicrobial susceptibility patterns of diarrheic E. coli and other food and waterborne pathogens
All the pathogens from the different sources were resistant to trimethoprim-sulfamethoxazole. Overall, amongst the 20 diarrheic E. coli isolates (used as sentinels for antimicrobial resistance surveillance), there was resistance to the following antibiotics: ampicillin, amoxicillin-clavulanate, cefoxitin, cefuroxime, cefotaxime, cefepime, ciprofloxacin, trimethoprim-sulfamethoxazole, and nitrofurantoin (Table 8).
Only three isolates were multi-drug resistant and were isolated from meat (one isolate) and stool (two isolates). All Plesiomonas (from water and stool) and Aeromonas (from meat and stool) were resistant to ampicillin while all Salmonella enterica Idikan and Salmonella enterica non – typeable were resistant to ciprofloxacin (Table 9).
Discussion
This study revealed a high prevalence of food and waterborne, as well as sanitation associated bacterial pathogens in meat, surface water and diarrheal stool from a low-income informal settlement setting in Cape Town, South Africa. Assessment of non-human sources for diarrheic bacterial pathogens is important because environmental modifications of potential reservoirs would aid the prevention of infections with drug-resistant pathogens.
Among the children that presented with diarrhoea, 5% (3/66), 20% (13/66), and 17% (11/33) reported bloody stool, vomiting, and fever respectively. Among the bacterial pathogens detected, S. flexneri, C. jejuni, STEC, and EIEC have been commonly reported to cause invasive disease. The associations between the presence of these bacterial pathogens and the different clinical presentations could not be determined due to small sample size. Notably, none (0/11) of the S. flexneri cases presented with bloody stool - a finding consistent with previous studies that reported the poor sensitivity of this clinical sign for the recovery of Shigellae [9, 10]. S. flexneri was the second most prevalent pathogen (the highest was DAEC) in this setting; it was the most prevalent among systemic shigellosis cases diagnosed between 2003 and 2009 in South Africa [26]. Shigella and other dysentery-causing infections may be underappreciated in primary health care in South Africa. Appropriate laboratory testing and surveillance to support the differential diagnosis and targeted treatment of enteric infections in vulnerable communities are urgently needed to aid patient care and reduce secondary transmission in the community. At least seven outbreaks of shigellosis or bacillary dysentery have been reported from South Africa since 1994. These have been associated with poor water and sanitation infrastructure. Our study community experienced an outbreak of bacillary dysentery in 1999 that affected 82 people and resulted in 3 deaths in children under two years of age [27]. Without access to laboratory testing, many cases will go undetected hence resulting in poor patient outcomes and increased health expenditure.
Similar studies in South Africa, albeit conducted in non – informal settlement settings, have reported a prevalence of 10% for Campylobacter (compared to 5% in this study) in the stool in the Venda region, Limpopo, South Africa [28]. Among the diarrheic E. coli that were detected in stool in this setting, DAEC were the most common (12%, 8/66 for isolates and 23%, 35/66 for marker genes) and EIEC were the least common (isolates 2%, 1/66 and marker genes 6%, 4/66). An earlier report in Egypt, 2013, established presence of DAEC associated with diarrhoea in human and non-human sources [29]. Our study highlights DAEC as a diarrhoea associated foodborne pathogen in this informal settlement setting. Even though EIEC is thought to be endemic in Africa settings causing traveller’s diarrhoea, our findings have shown that it was not prevalent in this local population. For all the diarrheic E. coli that were detected, more of the virulence marker genes were detected by these methods, compared to the isolates on CHROMagar™STEC from the same sample. Infection with multiple organisms was common (17% - 6/36 of the stool samples that carried a bacterial pathogen). All co-infected patients were infected with at least one of S. flexneri or DAEC. We did not test for infection or co-infection with viruses or intestinal parasites. Consistent with the reported inadequate and low second rotavirus vaccine coverage in the Cape Town metro [6] only 39% (26/66) of our cohort reported receiving rotavirus vaccination.
Most of the cases (79% - 52/66) used the communal standpipe as the primary source of water. According to the City of Cape Town Water and Sanitation Department, the communal standpipe nozzles in the informal settlements often were contaminated [30]. Lack of access to clean water is linked to high incidences of diarrheal disease [31].. Faecal contamination was demonstrated by the persistent significantly high levels of faecal coliforms in the local water canal that is accessible to domestic animals and humans. The increased level of contamination seen in the canal during the hottest and driest month of February (zero precipitation) is informative to aid waterborne disease risk management and public information, particularly during a period when human and animal exposure is more likely. The recovery of viable S. flexneri from open surface water in the community is alarming as Shigellae are particularly evanescent outside the host, suggesting a substantial presence. P. shigelloides and V. vulnificus were abundant in canal water raising concern for those accessing the canal for recreation, washing, food extraction or drinking; and to those executing monitoring activities. The prevalence of stx1 genes in water (30% of samples) was higher than the 15% previously detected in the Berg River system [32]. The presence of rfbE and wbdl in the water highlights it as a potential reservoir of STEC O157: H7 and STEC O111. This is of clinical and epidemiological significance because these are highly pathogenic STEC serotypes [33].
Raw meat samples collected from informal street-side vendors showed C. jejuni and S. enterica Idikan were most common pathogens identified. Of concern, A. sobria and P. shigelloides, typically found in aquatic habitats, were recovered from meats, raising concern over vendor access to safe water and sanitation infrastructure. S. enterica Idikan was isolated from 5% (4/85) of the meat samples. It has previously been isolated from food-producing animals; animal feeds, meat from abattoirs, water, and associated environments in South Africa at a prevalence of 6% (between 2012 and 2014) [34]. C. jejuni in South Africa is commonly isolated from poultry and is increasingly becoming resistant to antibiotics [35,36,37]. The microbiological standards for foodstuffs sold in South Africa regard the recovery of Salmonella, Shigella or E. coli of any quantity as unacceptable [38].
Considering diarrheic E. coli as sentinels for surveillance of foodborne antimicrobial resistance in this setting, these pathogens showed resistance to antibiotics usually used as first-line options for treatment of infections caused by Gram negative bacilli, such as ampicillin, amoxicillin-clavulanate, and extended-spectrum cephalosporins.
Limitations of this study
Pathogen-specific enrichment methods would have been more sensitive for isolate recovery. Viral and parasitic causes of diarrhoea were not targeted in this study.
Conclusions
We showed that antimicrobial resistance to a number of antimicrobials is present in diarrheal pathogens from human, food, and environmental sources in an informal settlement in Cape Town.S. flexneri spreads from person-to-person as a result of poor sanitation and inadequate quantities of water for personal hygiene, highlighting the need to urgently address inadequate water and waste provision in communities such as this one.
Globally, informal settlements present unique public health challenges due to dense human populations, poor residential infrastructure, and poor hygiene practices. Informal abattoirs do not follow recommended slaughter practice and often contaminate the environment. In a setting where diarrhoea is often empirically managed, knowledge of the sources, prevalence and antibiotic susceptibility patterns of bacteria causing enteric infections is critical.
Availability of data and materials
All data generated or analysed during this study are included in this published article. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- aat: Anti:
-
aggregation protein transporter gene
- AMK:
-
Amikacin
- ATCC:
-
American Type Culture Collection
- AZT:
-
Aztreonam
- bfp :
-
Bundle forming pilus
- CAZ:
-
Ceftazidime
- CDC:
-
Centre for Disease Control and Prevention
- CIN:
-
Gentamicin
- CIP:
-
Ciprofloxacin
- CLSI:
-
Clinical Laboratory Standards Institute
- COL:
-
Colistin
- CTX:
-
Cefotaxime
- daaC :
-
Fimbrial adhesion gene
- DAEC:
-
Diffusely adherent Escherichia coli
- DOR:
-
Doripenem
- DOX:
-
Doxycyclin
- eae :
-
intimin gene
- EAggEC:
-
Enteroaggregative Escherichia coli
- EIEC:
-
Enteroinvasive Escherichia coli
- EPEC:
-
Enteropathogenic Escherichia coli
- ETEC:
-
Enterotoxigenic Escherichia coli
- ETP:
-
Ertapenem
- F:
-
Female
- FEP:
-
Cefepime
- HUS:
-
Haemolytic Uremic Syndrome
- IMP:
-
Imipenem
- ipa :
-
invasive plasmid antigen
- LEV:
-
Levofloxacin
- LT:
-
Heat labile enterotoxin gene
- M:
-
Male
- MEM:
-
Meropenem
- MIC:
-
Minimum inhibitory concentration
- MIN:
-
Minocycline
- NHLS:
-
National Health Laboratory Services
- PCR:
-
Polymerase chain reaction
- POL:
-
Polymixin
- ST:
-
Heat stable enterotoxin gene
- STEC:
-
Shiga toxin producing Escherichia coli
- Stx:
-
Shiga toxin gene
- SXT:
-
Trimethoprim sulfamethoxazole
- Ter:
-
Tellurium resistance gene
- TGC:
-
Tigecycline
- TIM:
-
Ticarcillin clavulanic acid
- TOB:
-
Tobramycin
- TSB:
-
Tryptic soy broth
- TZP:
-
Piperacillin-Tazobactam
References
Sweijd, N. A., Wright C. Y., Westwood A. et al: Climate change is catchy – but when will it really hurt? SAMJ 2015, 105(12).
Dorrington RE, B. D., Laubscher R, Nannan N: rapid mortality surveillance report 2015. Cape Town: South African Medical Research Council; 2016.
Groenewald P, M. W., Morden E, Zinyakatira N, Neethling I, Daniels J, Evans J, Cornelius K, Berteler M, Martin LJ, Dempers J, Thompson V, Vismer M, Coetzee D, Bradshaw D. : Western cape mortality profile 2011. . Cape Town: South African Medical Research Council; 2014.
Lehohla, P.: South Africa Demographic and Health survey. In: key Indicators. Pretoria, South Africa, and Rockville, Maryland, USA. : National Department of Health, Statistics South Africa, South African Medical Research Council, and ICF; 2016: 25.
Msemburi W, P., Dorrington RE, Neethling I, Nannan N, Groenewald P, Laubscher R, Joubert J, Matzopoulos R, Nicol E, Nojilana B, Prinsloo M, Sithole N, Somdyala N and Bradshaw D. : Second national burden of disease study for South Africa. . In: Cause-of-death profile for South Africa, 1997–2012. Cape Town: South African Medical Research Council; 2016: 8.
N. McKerrow, M. M., G. Boon, A. Ferris: 2nd Triennial report of the committee on morbidity and mortality in children under five years. In.: Ministry of Health, South Africa.; 2014: 27.
Sengwana J. Diarrhoea management in primary health care facilities in the cape Metropole region. Mini-thesis: University of Western Cape; 2003.
The National Department of Health. Standard treatment guidelines and essential medicines list for South Africa: primary health care level. 5th ed. Pretoria, South Africa: The National Department of Health; 2014.
Kotloff KL, Nataro JP, Blackwelder WC, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the global enteric multicenter study, GEMS): a prospective, case-control study. Lancet. 2013;382(9888):209–22.
Keddy KH, Smith AM, Page NA. GEMS extend understanding of childhood diarrhoea. Lancet. 2016;388(10051):1252–4.
Govender T, Barnes JM, Pieper CH. Living in low-cost housing settlements in cape town, South Africa-the epidemiological characteristics associated with increased health vulnerability. Journal of urban health : bulletin of the New York Academy of Medicine. 2010;87(6):899–911.
http://www.statssa.gov.za/?page_id=4286&id=321 [http://www.statssa.gov.za/?page_id=4286&id=321].
World Health Organisation: Integrated Surveillance of Antimicrobial Resistance. In: Integrated Surveillance of Antimicrobial Resistance. Geneva, Switzerland: World Health Organisation; 2013: 8–10.
Environmental Protection Agency: Surface Water field sampling manual. In: Water column chemistry, bacteria, and flows. Ohio: Environmental Protection Agency; 2013: 13–24.
Ojo OE, Ajuwape AT, Otesile EB, et al. Potentially zoonotic Shiga toxin-producing Escherichia coli serogroups in the faeces and meat of food-producing animals in Ibadan Nigeria. Int J Food Microbiol. 2010;142(1–2):214–21.
07.100.200: Microbiology of Water. [http://www.iso.org/iso/iso_catalogue/catalogue_ics/catalogue_ics_browse.htm?ICS1=07&ICS2=100&ICS3=20].
Grys TE, Sloan LM, Rosenblatt JE, et al. Rapid and sensitive detection of Shiga toxin-producing Escherichia coli from nonenriched stool specimens by real-time PCR in comparison to enzyme immunoassay and culture. J Clin Microbiol. 2009;47(7):2008–12.
Kalule, J. B., Keddy K. H., Smith A. et al: Development of a real-time PCR assay and comparison to CHROMagarTM STEC to screen for Shiga toxin-producing Escherichia coli in stool, Cape Town, South Africa. AJLM 2017, 6(1).
Tau, N.P: Escherichia coli O104 associated with human diarrhoea, South Africa,2004-2011. EID 2012, 18(8).
Stacy-Phipps S, Mecca JJ, Weiss JB. Multiplex PCR assay and simple preparation method for stool specimens detect enterotoxigenic Escherichia coli DNA during course of infection. J Clin Microbiol. 1995;33(5):1054–9.
Vidal M, Kruger E, Durán C, et al. Single multiplex PCR assay to identify simultaneously the six categories of diarrheagenic Escherichia coli associated with enteric infections. J Clin Microbiol. 2005;43(10):5362–5.
Cebula TA, Payne WL, Feng P. Simultaneous identification of strains of Escherichia coli serotype O157:H7 and their Shiga-like toxin type by mismatch amplification mutation assay-multiplex PCR. J Clin Microbiol. 1995;33(1):248–50.
Tau NP, Meidany P, Smith AM, et al. Escherichia coli O104 associated with human diarrhea, South Africa, 2004-2011. Emerg Infect Dis. 2012;18(8):1314–7.
Paton AW, Paton JC. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J Clin Microbiol. 1998;36(2):598–602.
Bettelheim KA, Thompson CJ. New method of serotyping Escherichia coli: implementation and verification. J Clin Microbiol. 1987;25(5):781–6.
Keddy KH, Sooka A, Crowther-Gibson P, et al. Systemic shigellosis in South Africa. Clin Infect Dis. 2012;54(10):1448–54.
International Society for Infectious Disease: Dysentry outbreak in Nyanga. In., 1994 edn. Promed. mail archives: Cape Argus.; 1994.
Samie A, Obi CL, Barrett LJ, et al. Prevalence of Campylobacter species, Helicobacter pylori and Arcobacter species in stool samples from the Venda region, Limpopo, South Africa: studies using molecular diagnostic methods. J Inf Secur. 2007;54(6):558–66.
Shabana II, Zaraket H, Suzuki H. Molecular studies on diarrhea-associated Escherichia coli isolated from humans and animals in Egypt. Vet Microbiol. 2013;167(3–4):532–9.
City of Cape Town Water and Sanitation Department: Annual Report 2012/13. In: Annual Report 2012/13. Edited by City of Cape Town WaSD. City of Cape Town website: City of Cape Town; 2012/13: 45–47.
Hirai M, Roess A, Huang C, et al. Exploring geographic distributions of high-risk water, sanitation, and hygiene practices and their association with child diarrhea in Uganda. Glob Health Action. 2016;9(1):32833.
Ndlovu T, Le Roux M, Khan W, et al. Co-detection of virulent Escherichia coli genes in surface water sources. PLoS One. 2015;10(2):e0116808.
Johnson KE, Thorpe CM, Sears CL. The emerging clinical importance of non-O157 Shiga toxin-producing Escherichia coli. Clin Infect Dis. 2006;43(12):1587–95.
Magwedere, K., Rauff D., De Klerk G. et al: Incidence of Nontyphoidal Salmonella in Food-Producing Animals, Animal Feed, and the Associated Environment in South Africa, 2012–2014. Clinical Infectious Diseases 2015, 61(suppl_4):S283-S289.
Bester LA, Essack SY. Prevalence of antibiotic resistance in Campylobacter isolates from commercial poultry suppliers in KwaZulu-Natal South Africa. J Antimicrob Chemother. 2008;62(6):1298–300.
Bester LA, Essack SY. Observational study of the prevalence and antibiotic resistance of Campylobacter spp. from different poultry production systems in KwaZulu-Natal, South Africa. J Food Prot. 2012;75(1):154–9.
Bok HE, Greeff AS, Crewe-Brown HH. Incidence of toxigenic Campylobacter strains in South Africa. J Clin Microbiol. 1991;29(6):1262–4.
South African Bureau of Standards: Regulations governing the microbiological standards of foodstuffs in South Africa. In. Cape Town: SABS; 2017.
Acknowledgements
We acknowledge the Centre for Enteric Diseases of the National Institute for Infectious Disease, for serotyping the diarrheic E. coli. We thank the Cape Town Water and Sanitation Department for facilitating water testing.
Funding
This study was funded by the National Research Foundation, South Africa, RISE ACP scholarship and the AfDB-HEST program. The funding bodies had no role in the design of the study, collection, analysis, interpretation of data, and in writing of the manuscript.
Author information
Authors and Affiliations
Contributions
JBK and LR conceptualized the study and developed the research protocols. JBK conducted laboratory testing and participated in the writing of the manuscript. LR participated in supervision, data analysis, and manuscript preparation. MPN supervised JBK and participated in the writing of the manuscript. MV facilitated and provided valuable input and analysis of water testing. AS and NPT participated in the laboratory analysis of the isolates. KHK supervised the research activities, participated in data analysis and manuscript preparation. KHK was supervisor to JBK. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Written consent was obtained from all study participants or their guardians (for those under the age of 18 years). Ethics approval to conduct this study was obtained from Higher Research and Ethics Committee of the University of Cape Town, Faculty of Health Sciences (HREC 014/2015).
Research was conducted in compliance with the Helsinki declaration.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Kalule, J.B., Smith, A.M., Vulindhlu, M. et al. Prevalence and antibiotic susceptibility patterns of enteric bacterial pathogens in human and non-human sources in an urban informal settlement in Cape Town, South Africa. BMC Microbiol 19, 244 (2019). https://doi.org/10.1186/s12866-019-1620-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-019-1620-6