Abstract
Background
The transition to a parasitic lifestyle entails comprehensive changes to the selective regime. In parasites, genes encoding for traits that facilitate host detection, exploitation and transmission should be under selection. Slavemaking ants are social parasites that exploit the altruistic behaviour of their hosts by stealing heterospecific host brood during raids, which afterwards serve as slaves in slavemaker nests. Here we search for evidence of selection in the transcriptomes of three slavemaker species and three closely related hosts. We expected selection on genes underlying recognition and raiding or defense behaviour. Analyses of selective forces in species with a slavemaker or host lifestyle allowed investigation into whether or not repeated instances of slavemaker evolution share the same genetic basis.
To investigate the genetic basis of host-slavemaker co-evolution, we created orthologous clusters from transcriptome sequences of six Temnothorax ant species - three slavemakers and three hosts - to identify genes with signatures of selection. We further tested for functional enrichment in selected genes from slavemakers and hosts respectively and investigated which pathways the according genes belong to.
Results
Our phylogenetic analysis, based on more than 5000 ortholog sequences, revealed sister species status for two slavemakers as well as two hosts, contradicting a previous phylogeny based on mtDNA. We identified 309 genes with signs of positive selection on branches leading to slavemakers and 161 leading to hosts. Among these were genes potentially involved in cuticular hydrocarbon synthesis, thus species recognition, and circadian clock functionality possibly explaining the different activity patterns of slavemakers and hosts. There was little overlap of genes with signatures of positive selection among species, which are involved in numerous different functions and different pathways.
Conclusions
We identified different genes, functions and pathways under positive selection in each species. These results point to species-specific adaptations rather than convergent trajectories during the evolution of the slavemaker and host lifestyles suggesting that the evolution of parasitism, even in closely related species, may be achieved in diverse ways.
Similar content being viewed by others
Background
Parasitism is one of the most successful modes of life, as measured by how often it evolved and by how many parasitic species presently exist [1]. Parasites are a taxonomically highly diverse group, and range from intragenomic ‘genetic’ parasites, through microparasites (viruses, bacteria and protozoa) and macroparasites (worms, arthropods and even vertebrates) [2], to brood and social parasites [3, 4].
Parasites are ideal biological models for the study of ecological specialization, speciation mechanisms, diversification and co-adaptation [5]. The relationship of hosts and parasites is one of mutual adaptation with parasites trying to dupe the host, whereas hosts adapt to defend themselves [6,7,8,9,10,11,12]. Co-evolutionary dynamics are shaped by numerous factors including life history traits [13, 14], epidemiological characteristics [15, 16], population size [17, 18], fluctuating environmental changes [19], the presence of multiple parasites reviewed by [20], and social interactions within the host taxon reviewed by [21, 22].
Signatures of balancing selection are expected on immunity genes, playing a major role in the co-evolution between micro-parasites and their hosts, whereas genes encoding behavioural or morphological traits, important in social parasites and their hosts, should show signs of positive selection [23].
Social parasitism, is a special form of parasitism, where the social behaviour of the host, rather than its physiology, is exploited [3]. Avian brood parasites, such as cuckoos and cowbirds take advantage of the brood care behaviour of other bird species, and thus avoid the costs of parental care [24, 25]. Several avian brood parasites evolved from non-parasitic ancestors, and started out by exploiting the brood care behaviour of their conspecifics [26]. Similarly, ant social parasites, can arise via sympatric speciation from their later host [27]. Such a transition to a parasitic lifestyle should lead to the selection of traits important for a parasitic mode of life, such as host recognition, circumventing the host defence system, and transmission. Indeed, avian brood parasites lost their ability to build nests [28], a social parasitic wasp needs a specific host species to be successful as parasite [29], and many slavemaking ants are unable to even feed themselves and completely rely on the care of their enslaved host workers [30].
We are just starting to understand the genomic basis of the parasite lifestyle as such [31,32,33 and authors therein], and some first patterns of convergence, gene losses and gains become apparent [33 and authors therein, 34]. First studies on the genetic basis of social parasitism concentrated either on ant inquilines - social parasites, that secondarily lost the worker caste and inquiline queens live within host colonies [35] or the Cape honeybee (Apis mellifera capensis), in which workers invade other colonies and reproduce clonally [36]. Identified candidate loci for this form of social parasitism include genes involved in ecdysteroid signalling, juvenile hormone and dopamine biosynthesis, which may regulate worker ovary activation [37]. A study on three workerless inquiline social parasites of Vollenhovia ants in comparison to their Pogonomyrmex hosts found little evidence for gene loss, damaging mutations, or shifts in selection regimes, suggesting that regulatory changes, rather than sequence differences play a role in the evolution of these workerless social parasites [35]. However, the genomic basis of the slavemaker lifestyle and its’ peculiarities has never been investigated.
Here we explore the evolution of the slavemaker lifestyle in North American Temnothorax ants, a taxon in which slavery evolved several times independently [38]. We specifically focus on three slavemaker species T. americanus, T. duloticus and T. pilagens, and their three closely related host species, T. longispinosus, T. curvispinosus and T. ambiguus [39, 40]. The dulotic lifestyle of these three slavemakers is characterized by recurrent and destructive slave raids during summer [39]. During these raids, slavemaker worker raiding parties search for and attack host colonies to steal worker brood. Upon their emergence as adult workers in slavemaker nests, the social behaviours of these enslaved host workers will be exploited by the slavemakers, whose workers lost the ability to care for themselves [41]. While host nests on average contain around 50 workers [42], the number of workers in slavemaker nests is much lower with on average approximately five workers [39, 40, 43]. Moreover, slavemaker workers are only active during the raiding season and do not take over normal worker chores such as brood care and foraging [44, 45]. Each slavemaker species exhibits distinct morphological characteristics (e.g. size and colour), and raiding behaviours [41, 44]. T. americanus - the most derived parasite in the group in terms of morphology and behaviour - mainly uses a propaganda pheromone to induce panic among hosts, preventing organized evacuation or nest defence [41, 46, 47]. The strategy of T. pilagens is quite variable, and may also depend on the aggressiveness of the host colony [40, 48]. In some instances host workers are killed by stinging, while in other cases the raid is seemingly peaceful without any casualties, facilitating the incorporation of even adult host workers into the slavemaker colony [48]. T. duloticus is a fierce slavemaker that mostly stings all opponents to death before taking the brood, resulting in the local eradication of host colonies [41, 43, 44, 49]. Each of the three slavemakers can exploit several host species, but has a clearly preferred host. The derived T. americanus uses all three Temnothorax species, but focusses when possible on T. longispinosus [50]. T. duloticus occasionally attacks T. longispinosus but prefers T. curvispinosus [43] and T. pilagens prefers T. ambiguus over T. longispinosus [40, 48].
Co-evolution between the obligate social parasites and their hosts not only leads to adaptations in slavemakers, but also to counter-adaptations in behavioural, chemical and life history traits in host species and populations [11, 47, 51,52,53,54]. Host aggression [54], as well as host defence strategies [55] are linked to geographic variation in parasite pressure. It is known that adaptations to similar ecological conditions may lead to the evolution of similar (convergent) phenotypes in non-related species. The degree to which parallelism extents to the molecular level has recently experienced an upsurge of interest [56,57,58,59,60]. Evidence is ambiguous, with some studies pointing to parallelism, and others to species-specific trajectories [59, 61 and authors therein]. Moreover, it becomes clear that the level of organisation plays a major role in detecting convergent evolution, as the degree of parallelism is predicted to increase from the nucleotide level to features of whole organisms [61]. The North American Temnothorax system, with six closely related slavemaker and host species is ideal to study the genetic basis of repeated evolution of phenotypic traits involved in host-parasite co-evolution.
The main objective of this study was to investigate the selective forces shaping the host and slavemaker lifestyles, and the organisational level of convergence. The main questions we tried to answer were: Which genes are under positive selection in slavemakers or hosts? Is molecular parallelism involved in the convergent evolution of slavemaker lifestyles? Do we find convergence on the gene, functional or pathway level?
Methods
Ant colonies were collected over the course of 2 years (2013 and 2014) in New York State, Ohio and Michigan (Additional file 1), and brought back to the lab in Mainz. To induce raiding activity, colonies were moved during the raiding season in August to 25 °C, 14 L:10D light cycle conditions 1 week prior to the onset of the raiding experiment. Raiding arenas (30 × 40 cm plastic boxes with plastered floor) were set up, in which each slavemaker species was allowed to raid colonies of its preferred host species. We waited until slavemaker scouts had returned to their mother nest and recruited additional raiders to infiltrate the host nest, and aggressive encounters between slavemaker and host workers could be observed. This was the time we sampled workers actively engaging in a raid or nest defence respectively. To obtain workers in a somewhat neutral behavioural state, we collected host individuals outside the nest before raids, as well as slavemaker workers outside the raiding season under the same external conditions. Since we were interested in the evolution of slavemaker and host genes in respect to raiding and nest defence behaviour, we obtained transcriptomes of ants engaged in these respective behaviours. Six workers per species and behaviour were pooled for RNA isolation in replicates of six. Libraries were constructed and sequenced paired-end on an Illumina HiSeq 2000 at GENterprise Genomics. Sequences were quality trimmed with Trimmomatic v0.32 [62]. De novo assembly of the transcriptomes was conducted using a combination of the CLC bio workbench (Qiagen) and MIRA [63] (Additional file 2; for more details see [64]). Contigs were annotated using BlastX v.2.2.30 against the non-redundant arthropod database (November 2014). The online tool ORFpredictor 2.0.3 [65] was used to predict open reading frames and amino acid sequences for all contigs. The predicted and translated amino acid sequences were used as input for OrthoMCL 2.0.9 [66], to build ortholog sequence clusters. In total we obtained 55,521 orthologous protein clusters, out of which 6432 clusters contained at least one sequence per species. These clusters were filtered with an in-house python script (available from GitHub: https://zenodo.org/record/60135#. V9k495h96Uk) based on pairwise Blast similarity scores, which resulted in 5791 clusters with a single sequence per species. After trimming these sequence alignments with Gblocks 0.91b [67], 5199 clusters remained for further analyses (NCBI Bio Project GSE95604).
Phylogenetic analysis
We chose the myrmicine ant Acromyrmex echinator to include as outgroup, for which we observed the highest Blast similarity in our contig Blast searches. A. echinator sequences were obtained from the “Hymenoptera Genome Database” [68; aech_OGSv3.8_pep.fa]. We inferred orthology between T. curvispinosus and Acromyrmex echinator applying a local BLASTn [69]. The according sequences for each cluster were obtained and aligned with Mafft 7.0 [70]. The alignments were trimmed using Gblocks 0.91b [66] with default settings. All clusters were concatenated into a single alignment and the program ProtTest 3.4 [71] was used to calculate the appropriate evolutionary model (JTT + I + G + F). A Maximum Likelihood phylogenetic tree with 1000 bootstrap replicates was constructed with RAxML 8.1.16 [72]. We additionally estimated evolutionary models for each single cluster and constructed the respective Maximum Likelihood trees for the codeml analyses (see below).
Tests for positive and relaxed selection
To test for signatures of positive selection the software package PAML 4.8 [73] was used to apply the branch-site model A in codeml (model = 2, NSsites = 2). codeml estimates the nonsynonymous/synonymous substitution ratio (ɷ = dN/dS), where ɷ = 1 indicates neutral evolution, ɷ < 1 purifying selection, and ɷ > 1 indicates positive selection. To test for statistical significance log-likelihood ratios were calculated and FDR corrected for multiple testing [74]. The cluster specific tree topology, as inferred by RAxML was used as input for codeml. To test for positively selected genes, we coded each single species as foreground branch, and additionally the set of slavemaker branches as well as the host branches respectively.
The online tool Venny 2.1 (http://bioinfogp.cnb.csic.es/tools/venny/) was used to visualize shared and species-specific genes. To statistically assess to what extent the observed intersection in divergent features among pairs of species would be expected by chance, we applied a randomisation procedure implemented in a custom Python script. We used 10,000 replicates to infer how often an observed intersection of size i of x and y positive draws from a base population of size z was larger or smaller than those from random draws.
Enrichment analyses
To obtain identifiers suitable for the enrichment tool DAVID 6.7 [75], we inferred orthology by applying a BLASTx between T. curvispinosus contigs and Drosophila melanogaster protein sequences (dmel-all-translation-r5.56.fasta) obtained from flybase (flybase.org). The complete contig set was used as background and the according positively selected genes as test set. Furthermore, to obtain pathway information in form of KO (KEGG Orthology) assignments for the according gene sets, we utilized KAAS [76], an automated annotation server. The KEGG Mapper – Reconstruct Pathway tool was used to obtain the associated pathways (http://www.genome.jp/kegg/tool/map_pathway.html).
Results
Phylogenetic analysis
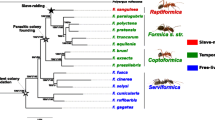
Based on the concatenated sequence of 5199 ortholog sequence clusters the RAxML phylogenetic analysis resulted in a tree with well supported nodes (Fig. 1). T. americanus is the most distant taxa to the other five Temnothorax species, as corroborated by its deviant phenotype and a previous phylogenetic analysis [38]. In contrast to this earlier phylogeny based on two mitochondrial loci [38], our nuclear tree now supports a sister species relationship between the two younger slavemaking species T. duloticus and T. pilagens [40], as well as between the two host species T. ambiguus and T. longispinosus, with T. curvispinosus being the next distant taxon, followed by T. americanus.
RAxML obtained Maximum Likelihood phylogenetic relationship of the six slavemaker and host species with Acromyrmex echinatior as outgroup, and based on 5199 ortholog gene-clusters (ML bootstrap percentages depicted at nodes). Slavemaker species names are given in red, host species names in black. Arrows connect slavemaker-host species pairs
Positively selected genes
In total, we found 574 genes under positive selection; 309 on the branches leading to slavemakers, and significantly less, 161, positively selected genes on the branches to hosts (χ2 1 = 77.85, p < 0.0001; Additional file 3). Looking at the branches of each single species, we detected more than four times as many genes under selection in the derived slavemaker T. americanus (N = 211) than in the two younger slavemaker sister species T. pilagens (N = 38) and T. duloticus (N = 54; Fig. 2a). The host species T. ambiguus shares one gene under positive selection with its sister species T. longispinosus, and two genes with T. curvispinosus, whereas the latter two species do not share any positively selected genes (Fig. 2b). In slavemakers, one positively selected gene (a hypothetical protein) is shared amongst all three species, T. americanus shares two genes each with the two other slavemakers, and T. pilagens shares only one positively selected gene with its sister T. duloticus (Additional file 3). In both, hosts and slavemakers, the number of shared genes under positive selection among species is less than one would expect by chance (Additional file 4: Table S1). Moreover, we compared the positively selected genes to differentially expressed genes from an accompanying gene expression analysis [64] based on the same host and slavemaker transcriptomes. Six genes in hosts and 36 in slavemakers appeared in both, the differential expression and the positive selection analyses (Table 1). None of the positively selected gene sets per species, in slavemakers or hosts, were enriched for any functional category.
Venn diagrams depicting the number of shared and “private” positive selected genes (a + b) and corresponding pathways (c + d) in slavemakers and hosts. (T. dul = T. duloticus, T. ameri = T. americanus, T. pila = T. pilagens, T. curvi = T. curvispinosus, T. longi = T. longispinosus, T. ambi = T. ambiguus)
The comparison of pathways associated with selected genes indicates that selected genes between species not only belong to different functional categories, but also to many different pathways (Additional file 5). In slavemakers, we identified 128 different pathways amongst the positively selected genes, the majority (77%) of which were also species-specific (χ2 1 = 73.26, p < 0.001; Fig. 2c). The genes positively selected in hosts belong to 102 different pathways, the majority (70%) of which were species-specific (χ2 1 = 29.82, p < 0.001; Fig. 2d). Nevertheless, more pathways were shared than expected by chance in hosts and also in slavemakers, except between the sister species T. pilagens and T. duloticus (Additional file 4: Table S2). The eight pathways shared among hosts are the metabolic pathway, biosynthesis of secondary metabolites, biosynthesis of antibiotics, p53 signalling, PI3K-Akt signalling, Wnt signalling, thyroid hormone signalling and the longevity regulating pathway. In slavemakers, four pathways were shared amongst all three species including metabolic pathways, biosynthesis of secondary metabolites, biosynthesis of antibiotics and endocytosis.
Slavemaker-host pairs
Genes under selection in slavemaker-host pairs should show little overlap due to their coevolution, as different traits are under selection in slavemakers and hosts. An exception could be cuticular hydrocarbon genes, when slavemakers try to mimic host profiles [77] and utilize the same genes as their closely related hosts. A more important cause of overlap in selected genes in slavemaker and host pairs might be, that both species inhabit the same habitat and therefore adapt to the same environmental conditions. We investigated the number and functions of positively selected genes and pathways between each slavemaker-host pair in order to make inferences on local adaptation. We found between none and four shared positively selected genes (Fig. 3a-c; Additional file 6), and 7–23 shared pathways between pairs (Fig. 3d-f). The number of shared genes and pathways was higher than expected in the pair including the most diverged parasite species T. americanus – T. longispinosus, and as expected by chance in the other two parasite-host pairs (Additional file 4: Table S3 and S4). Moreover, we tested whether slavemakers share more genes with their preferred host in contrast to the other host species. T. duloticus shares more genes with T. longispinosus (n = 5) in comparison to its preferred host T. curvispinosus (n = 0) (X2 1 = 4.811, p = 0.028). In all other cases, the number of shared genes between preferred host and the other species did not differ (results not shown).
Venn diagrams depicting shared and “private” selected genes (a-c) and pathways (d-f) between host and slavemakers species pairs. For species name abbreviations please refer to Fig. 2
Discussion
The slavemaker lifestyle evolved several times independently in ants, with a hotspot of slavery evolution in the genus Temnothorax [37]. As slavemakers and their hosts are engaged in a constant co-evolutionary arms-race, the evolution of ant slavery is tightly linked to the evolution of behaviour, physiology and morphology in their hosts [78,79,80]. The focus of our study was the genomic basis of the (co-) evolution of the slavemaker and host lifestyle. We were thus interested in identifying genes with signatures of positive selection in slavemakers and hosts respectively. Furthermore, we asked whether or not, and at which organisational level, the three slavemaker/host species show signs of genetic convergence; or whether each species follows its own specific evolutionary trajectory.
Positively selected genes
We identified twice as many genes under positive selection on branches leading to slavemakers as compared to host branches. This finding is in line with our expectation that the derived slavemaker mode of life should have led to the selection of more genes in comparison to the ancestral host lifestyle. However, based on the number of species-specific selected genes it becomes evident that 70% of the positively selected genes in slavemakers can be assigned to T. americanus only. All other slavemaker and host species have comparable and lower numbers of positively selected genes. T. americanus is the most distantly related species in this taxon and its behaviour and morphology are most derived from the other species. T. americanus workers have large square heads, which make them easily distinguishable from their hosts. They do not engage in normal worker behaviour, such as brood care or foraging, and are so dependent upon enslaved hosts that T. americanus will starve to death if not fed. During raids, they manipulate host behaviour via the release of glandular secretions [11, 41, 46], but never use their stinger, which is a typical behaviour for other Temnothorax hosts during aggressive interactions [7, 41, 48]. Foraging and brood care are standard behavioural repertoires in the hosts, and in the lack of slaves, will still be performed by T. duloticus [41, 44] and T. pilagens (pers. observation) slavemakers to some extent. In addition to many lifestyle differences, the longer evolutionary history with the possibility for co- and counter adaptations to T. longispinosus and its host ancestor, might explain the large number of positively selected genes in T. americanus in contrast to the other species.
Within the 309 positively selected genes on the branches to slavemakers, we were able to identify several candidate genes with a possible link to their slavemaker lifestyle. Amongst these, three different DNAJ-like protein subfamily members, which are heat shock protein homologs, and function as co-chaperons. They are involved in stress response in humans [81], and could thus play a role during stressful slave raids into fiercely defending host colonies.
Tachykinin is positively selected on the branches leading to hosts and might thus be a candidate gene for the host lifestyle. Tachykinin has been linked to aggressive behaviour in Drosophila [82 and authors therein], and recently also in ants [83]. Temnothorax hosts need to defend their nest aggressively, not only against intra-specific intruders, but particularly against slavemakers. Thus selection might specifically act on this gene in hosts. This is further corroborated by the fact that intra-specific aggression increases with the prevalence of the slavemaker T. americanus in the population [54].
In comparison to an accompanying gene expression analysis including the same six host and slavemaker sequence data [64], we found six genes to be both, differentially expressed and under positive selection in hosts and 36 in slavemakers. These genes are thus prime candidates for the evolution of the slavemaker and host lifestyle. Firstly, they are directly involved in raiding behaviour. Secondly, they show signs of positive selection. Among these genes Trypsin-7 was identified in two host species and the slavemaker T. americanus. Trypsin-7 is known for its function in digestion, e.g. it is blood meal induced in Anopheles gambiae, and may also play a role in host seeking behaviour [84]. It may thus be involved in host seeking behaviour in slavemakers and slavemaker detection in hosts. Endogenous daily (circadian) and annual (circannual) rhythms serve as biological clocks that provide the major basis for timing in most organisms [85]. Annual timing mechanisms regulate seasonal timing of reproduction, moult, and hibernation [86 and authors therein]. Positive selection on the circadian clock controlled protein, in slavemakers compared to hosts, suggests that this gene may regulate the aberrant activity patterns of slavemaker workers. Slavemakers are only active during raiding season in summer, and are taken care of for the rest of the year by the slaves [39, 87]. This changed activity pattern might thus be manifested by changes in the circadian rhythm. Two more genes of interest with possible direct link to the slavemaker evolution are “Elongation of very long chain fatty acids protein” in T. duloticus and Fatty acyl-CoA reductase 1 in T. pilagens. Both genes could be involved in the synthesis of cuticular hydrocarbons and thus might play a role in the avoidance of host recognition [88]. Indeed, a recent study on the cuticular hydrocarbon profiles of the same six species, revealed that slavemakers show consistently different chemical profiles than the three host species [89]. A recent switch to a parasitic lifestyle could thus have led to selection on genes underlying hydrocarbon synthesis.
In slavemakers only four pathways are shared among species. Three of these (metabolic pathways, biosynthesis of secondary metabolites, and biosynthesis of antibiotics) were also identified in the hosts, and only endocytosis is slavemaker specific. In hosts, the genes with signatures of positive selection belong to 102 different pathways, eight of which are shared among the three species. The PI3K-Akt pathway is part of the mTOR pathway regulating the cell cycle, and also known for its function in longevity [90]. Furthermore the longevity regulating pathway is shared among all three host species, reinforcing the importance of longevity within these host species. Despite their small body and colony size Temnothorax ants are quite long-lived with workers living up to a few years and queens over two decades [91]. Social Hymenopterans are known for a change in the longevity-fecundity trade-off with queens being both long-lived and highly fecund compared to the short-lived sterile workers [92,93,94,95,96,97]. We identified two pathways, which may play a role in morphological differences between the species and their adaptive divergence. The Wnt signalling pathway is known for its role in regulating key events during embryonic patterning and morphogenesis [98], and the thyroid hormone signalling pathway (in humans) is involved in the regulation of growth development and metabolism. The latter has been shown to play a role in the adaptive divergence of sticklebacks [99].
Slavemaker – Host pairs
Besides determining similarities in possible selection pressures within slavemakers and within hosts, we additionally investigated similarities in slavemaker-host pairs, because of their shared environment. We hypothesized that genes shared by both slavemakers and hosts with signatures of positive selection might give indication on local environmental selection pressures; though they could also represent genes involved in the co-evolutionary arms race. However, on the gene level there is hardly any congruence between slavemaker and host pairs (0–4 overlapping genes). On the pathway level, some of the above mentioned candidates appear, such as thyroid hormone signalling pathway in T. ambiguus and T. pilagens, PI3K-Akt and Wnt signalling pathway between T. americanus and T. longispinosus. In the T. duloticus – T. curvispinosus pair we identified circadian rhythm as well as the FoxO signalling pathway. Among others the latter coordinates the response to environmental changes, including metabolic stress (starvation) and oxidative stress [100]. Hence, these two pathways may give evidence for environmental selection pressures, e.g. temperature, seasonality, or food availability, experienced by T. duloticus and T. curvispinosus which co-occur in the same environment, in comparison to the other four species which are from different locales.
Conclusions
Our positively selected gene analyses revealed several candidate genes with a possible link to the slavemaker lifestyle, which are involved in the cuticular hydrocarbon profile composition, thus species recognition, and the aberrant activity pattern of slavemaker workers. To verify the functional and phenotypic importance of these candidates will now be the next step.
Furthermore, the results show little overlap of selected genes between species. On the pathway level however, we find higher congruence between species than expected, even though the majority of selected pathways remain species specific. Furthermore, the genes under positive selection belong to a wide variety of functions, as indicated by negative results in the enrichment analyses. The same pattern was identified in social parasitic cape honeybees [37]. It thus seems that the evolution of social parasites, including slavemakers is a broad encompassing process with species-specific evolutionary trajectories, based on selection in many genes with different functionality and pathway affiliation. Our results support the hypothesis that evolution is the unrepeatable result of stochastic events with highly contingent effects [101].
Abbreviations
- L:D:
-
Light: dark
- dN/dS:
-
Ratio of non-synonymous to synonymous mutations
- mtDNA:
-
Mitochondrial DNA
References
Poulin R, Morand S. The diversity of parasites. Q Rev Biol. 2000;75:277–93.
Viney M, Cable J. Macroparasite life histories. Curr Biol. 2011;21:R767–74.
Buschinger A: Bettel, Raub und Mord: aus dem Leben sozialparasitischer Ameisen. Verh Westdtsch Entomol 1993, 1993:7–20.
Davies NB, Bourke AFG, Brooke M de L. Cuckoos and parasitic ants: Interspecific brood parasitism as an evolutionary arms race. Trends Ecol Evol. 1989;4:274–8.
de Meeûs T, Michalakis Y, Renaud F. Santa Rosalia revisited: or why are there so many kinds of parasites in “the garden of earthly delights”? Parasitol Today. 1998;14:10–3.
Dawkins R, Krebs JR. Arms races between and within species. Proc R Soc Lond B Biol Sci. 1979;205:489–511.
Foitzik S, de Heer CJ, Hunjan DN, Herbers JM. Coevolution in host-parasite systems: behavioural strategies of slave-making ants and their hosts. Proc R Soc Lond B. 2001;268:1139–46.
Calcagno V, Dubosclard M, de Mazancourt C. Rapid exploiter-victim coevolution: the race is not always to the swift. Am Nat. 2010;176:198–211.
Kilner RM, Langmore NE. Cuckoos versus hosts in insects and birds: adaptations, counter-adaptations and outcomes. Biol Rev Camb Philos Soc. 2011;86:836–52.
Grasso DA, Mori A, Le Moli F. Analysis of the aggression between slave and slave-making (facultative and obligatory) ant species (Hymenoptera Formicidae). Ethology Ecol Evol. 1992;4:81–4.
Jongepier E, Foitzik S: Ant recognition cue diversity is higher in the presence of slavemaker ants. Behav Ecol. 2015;27:304–11.
Ortolani I, Zechini L, Turillazzi S, Cervo R. Recognition of a paper wasp social parasite by its host: evidence for a visual signal reducing host aggressiveness. Anim Behav. 2010;80:683–8.
Barrett LG, Thrall PH, Burdon JJ, Linde CC. Life history determines genetic structure and evolutionary potential of host–parasite interactions. Trends Ecol Evol. 2008;23:678–85.
Strauß JF, Crain P, Schulenburg H, Telschow A. Experimental evolution in silico: a custom-designed mathematical model for virulence evolution of Bacillus thuringiensis. Zool (Jena). 2016;119:359–65.
Tellier A, Brown JKM. Stability of genetic polymorphism in host–parasite interactions. Proc R Soc B Biol Sci. 2007;274:809–17.
González-Tortuero E, Rusek J, Turko P, Petrusek A, Maayan I, Piálek L, Tellenbach C, Gießler S, Spaak P, Wolinska J. Daphnia parasite dynamics across multiple Caullerya epidemics indicate selection against common parasite genotypes. Zoology. 2016;119:314–21.
Papkou A, Gokhale CS, Traulsen A, Schulenburg H. Host – parasite coevolution: why changing population size matters. Zoology. 2016;119:330–8.
Gandon S, Michalakis Y. Local adaptation, evolutionary potential and host-parasite coevolution: interactions between migration, mutation, population size and generation time. J Evol Biol. 2002;15:451–62.
Wolinska J, King KC. Environment can alter selection in host–parasite interactions. Trends Parasitol. 2009;25:236–44.
Bose J, Kloesener MH, Schulte RD. Multiple-genotype infections and their complex effect on virulence. Zoology. 2016;119:339–49.
Kurze C, Routtu J, Moritz RFA. Parasite resistance and tolerance in honeybees at the individual and social level. Zoology. 2016;119:290–7.
Joop G, Vilcinskas A. Coevolution of parasitic fungi and insect hosts. Zoology. 2016;119:350–8.
Croze M, Živković D, Stephan W, Hutter S. Balancing selection on immunity genes: review of the current literature and new analysis in Drosophila melanogaster. Zoology. 2016;119:322–9.
Brooke M, Davies NB. Egg mimicry by cuckoos Cuculus canorus in relation to discrimination by hosts. Nature. 1988;335:630–2.
Davies NB. Cuckoos, cowbirds and other cheats. London: T. & A. D. Poyser; 2000.
Yamauchi A. Theory of evolution of nest parasitism in birds. Am Nat. 1995;145:434–56.
Leppänen J, Seppä P, Vepsäläinen K, Savolainen R. Genetic divergence between the sympatric queen morphs of the ant Myrmica rubra. Mol Ecol. 2015;24:2463–76.
Hamilton WJ, Orians GH. Evolution of brood parasitism in altricial birds. Condor. 1965;67:361–82.
Fanelli D, Henshaw M, Cervo R, Turillazzi S, Queller DC, Strassmann JE. The social parasite wasp Polistes atrimandibularis does not form host races. J Evol. 2005;18:1362–7.
Darwin C. On the origin of species. Cambridge, Mass: Harvard University Press; 1859.
Zhang L, Zhou Z, Guo Q, Fokkens L, Miskei M, Pócsi I, Zhang W, Chen M, Wang L, Sun Y, Donzelli BGG, Gibson DM, Nelson DR, Luo J-G, Rep M, Liu H, Yang S, Wang J, Krasnoff SB, Xu Y, Molnár I, Lin M. Insights into adaptations to a near-obligate nematode endoparasitic lifestyle from the finished genome of Drechmeria coniospora. Sci Rep. 2016;6:23122.
Skippington E, Barkman TJ, Rice DW, Palmer JD. Miniaturized mitogenome of the parasitic plant Viscum scurruloideum is extremely divergent and dynamic and has lost all nad genes. PNAS. 2015:112:E3515–24.
Jackson AP. Preface: The evolution of parasite genomes and the origins of parasitism. Parasitology. 2015;142:S1–5.
Poulin R, Randhawa HS. Evolution of parasitism along convergent lines: from ecology to genomics. Parasitology. 2015;142:S6–S15.
Smith CR, Helms Cahan S, Kemena C, Brady G, Yang W, Bornberg E, Smith CR, Cahan SH, Kemena C, Eriksson T, Gadau J, Helmkampf M, Gotzek D, Okamoto M, Suarez AV, Mikheyev A. How do genomes create novel phenotypes ? Insights from the loss of the worker caste in ant social parasites. Mol Biol Evol. 2015;32:2919–31.
Goudie F, Allsopp MH, Solignac M, Beekman M, Oldroyd BP. The frequency of arrhenotoky in the normally thelytokous Apis mellifera capensis worker and the clone reproductive parasite. Insect Soc. 2015;62:325–33.
Wallberg A, Pirk CW, Allsopp MH, Webster MT. Identification of multiple loci associated with social parasitism in honeybees. PLoS Genet. 2016;12:1–30.
Beibl J, Stuart RJ, Heinze J, Foitzik S. Six origins of slavery in formicoxenine ants. Insect Soc. 2005;52:291–7.
Herbers JM, Foitzik S, Collins F. The ecology of slavemaking ants and their hosts in north temperate forests. Ecology. 2002;83:148–63.
Seifert B, Kleeberg I, Feldmeyer B, Pamminger T, Jongepier E, Foitzik S. Temnothorax pilagens sp. n. – a new slave-making species of the tribe Formicoxenini from North America (Hymenoptera, Formicidae). Zookeys. 2014;368:65–77.
Alloway TM. Raiding behaviour of two species of slave-making ants, Harpagoxenus americanus (Emery) and Leptothorax duloticus Wesson (Hymenoptera: Formicidae). Anim Behav. 1979;27:202–10.
Foitzik S, Achenbach A, Brandt M. Locally adapted social parasite affects density, social structure, and life history of its ant hosts. Ecology. 2009;90:1195–206.
Johnson CA, Herbers JM. Impact of parasite sympatry on the geographic mosaic of coevolution. Ecology. 2006;87:382–94.
Wilson EO. Leptothorax duloticus and the beginnings of slavery in ants. Evolution. 1975;29:108–19.
Stuart RJ, Alloway TM. Behavioural evolution and domestic degeneration in obligatory slave-making ants (Hymenoptera: Formicidae: Leptothoracini). Anim Behav. 1985;33:1080–8.
Brandt M, Heinze J, Schmitt T, Foitzik S. Convergent evolution of the Dufour’s gland secretion as a propaganda substance in the slave-making ant genera Protomognathus and Harpagoxenus. Insect Soc. 2006;53:291–9.
Jongepier E, Kleeberg I, Foitzik S. The ecological success of a social parasite increases with manipulation of collective host behaviour. J Evol Biol. 2015;12:2152–62.
Kleeberg I, Foitzik S. The placid slavemaker: avoiding detection and conflict as an alternative, peaceful raiding strategy. Behav Ecol Sociobiol. 2016;70:27–39.
Hare JF, Alloway TM. Prudent Protomognathus and despotic Leptothorax duloticus: differential costs of ant slavery. PNAS. 2001;98:12093–6.
Brandt M, Foitzik S. Community context and specialization influence coevolution between a slavemaking ant and its hosts. Ecology. 2004;85:2997–3009.
Pamminger T, Scharf I, Pennings PS, Foitzik S. Increased host aggression as an induced defense against slave-making ants. Behav Ecol. 2011;22:255–60.
Scharf I, Pamminger T, Foitzik S. Differential response of ant colonies to intruders: attack strategies correlate with potential threat. Ethology. 2011;117:731–9.
Kleeberg I, Pamminger T, Jongepier E, Papenhagen M, Foitzik S. Forewarned is forearmed: aggression and information use determine fitness costs of slave raids. Behav Ecol. 2014;25:1058–63.
Kleeberg I, Jongepier E, Job S, Foitzik S. Geographic variation in social parasite pressure predicts intraspecific but not interspecific aggressive responses in hosts of a slavemaking ant. Ethology. 2015;121:1–9.
Jongepier E, Kleeberg I, Job S, Foitzik S. Collective defence portfolios of ant hosts shift with social parasite pressure. Proc R Soc B. 2014;281:20140225.
Conte GL, Arnegard ME, Peichel CL, Schluter D. The probability of genetic parallelism and convergence in natural populations. Proc R Soc B. 2012;279:5039–47.
Martin A, Orgogozo V. The loci of repeated evolution: a catalog of genetic hotspots of phenotypic variation. Evolution. 2013;67:1235–50.
Elmer KR, Meyer A. Adaptation in the age of ecological genomics: insights from parallelism and convergence. Trends Ecol Evol. 2011;26:298–306.
Stern DL. The genetic causes of convergent evolution. Nat Rev Genet. 2013;14:751–64.
Soria-Carrasco V, Gompert Z, Comeault AA, Farkas TE, Parchman TL, Johnston JS, Buerkle CA, Feder JL, Bast J, Schwander T, Egan SP, Crespi BJ, Nosil P. Stick insect genomes reveal natural selection’s role in parallel speciation. Science. 2014;344:738–42.
Lobkovsky AE, Koonin EV. Replaying the tape of life: quantification of the predictability of evolution. Front Genet. 2012;3:246.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20.
Chevreux B, Wetter T, Suhai S. Genome sequence assembly using trace signals and additional sequence information. Comput Sci Biol Proc Ger Conf Bioinforma. 1999;99:45–56.
Alleman A, Feldmeyer B, Foitzik S. Comparative analyses of co-evolving host-parasite associations reveal unique expression patterns and pathways underlying slavemaker raiding and host defensive behavior in ants. in prep.
Min XJ, Butler G, Storms R, Tsang A. OrfPredictor: predicting protein-coding regions in EST-derived sequences. Nucleic Acids Res. 2005;33:W677–80.
Li L, Stoeckert CJ, Roos CJ. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–89.
Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–52.
Nygaard S, Zhang G, Schiøtt M, Nygaard S, Zhang G, Schiøtt M, Li C, Wurm Y, Hu H, Zhou J, Ji L, Qiu F, Rasmussen M, Pan H, Hauser F, Krogh A, Grimmelikhuijzen CJP, Wang J. The genome of the leaf-cutting ant Acromyrmex echinatior suggests key adaptations to advanced social life and fungus farming. Genome Res. 2011;21:1339–48.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10.
Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80.
Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–5.
Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–90.
Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–91.
Benjamni Y, Hochberg Y, Benjamini Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B-Methodological. 1995;57:289–300.
Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009:1–13.
Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–5.
Achenbach A, Witte V, Foitzik S. Brood exchange experiments and chemical analyses shed light on slave rebellion in ants. Behav Ecol. 2010;21:948–56.
Gross P. Insect behavioral and morphological defenses against parasitoids. Annu Rev Entomol. 1993;38:251–73.
Hart BL. Behavioural defence. In: Clayton DH, Moore J, editors. Host-parasite evolution. General principles and avian models. Oxford: Oxford University Press; 1997. p. 59–77.
Sorci G. Immunity, resistance and tolerance in bird – parasite interactions. Parasite Immunol. 2013;35:350–61.
Terada K, Mori M. Human DnaJ homologs dj2 and dj3, and bag-1 are positive cochaperones of hsc70. J Biol Chem. 2000;275:24728–34.
Pavlou HJ, Neville MC, Goodwin SF. Aggression: Tachykinin is all the rage. Curr Biol. 2014;24:R243–4.
Howe J, Schiøtt M, Boomsma JJ. Tachykinin expression levels correlate with caste-specific aggression in workers of the leaf-cutting ant Acromyrmex echinatior. Front Ecol Evol. 2016;4(May):1–12.
Muller HM, Catteruccia F, Vizioli J, Dellatorre A, Crisanti A. Constitutive and blood meal-induced trypsin genes in Anopheles gambiae. Exp Parasitol. 1995;81:371–85.
Gwinner E. Circadian and circannual programmes in avian migration. J Exp Biol. 1996;199:39–48.
Wikelski M, Martin LB, Scheuerlein A, Robinson MT, Robinson ND, Helm B, Hau M, Gwinner E. Avian circannual clocks: adaptive significance and possible involvement of energy turnover in their proximate control. Philos Trans R Soc Lond B. 2008;363:411–23.
Pohl S, Foitzik S. Slave-making ants prefer larger, better defended host colonies. Anim Behav. 2011;81:61–8.
Blomquist GJ, Bagnères A-G. Insect hydrocarbons biology, biochemistry and ecology. Cambridge, UK: Cambridge University Press; 2010.
Kleeberg I, Menzel F, Foitzik S. The influence of slavemaking lifestyle, caste and sex on chemical profiles in Temnothorax ants: insights into the evolution of cuticular hydrocarbons. Proc R Soc B. 2017;284:20162249.
Hay N. Interplay between FOXO, TOR, and Akt. Biochim Biophys Acta. 2011;1813:1965–70.
Keller L, Genoud M. Extraordinary lifespans in ants: a test of evolutionary theories of ageing. Nature. 1997;389:958–60.
Heinze J, Frohschammer S, Bernadou A. Queen life-span and total reproductive success are positively associated in the ant Cardiocondyla cf. kagutsuchi. Behav Ecol Sociobiol. 2013;67:1555–62.
Oettler J, Schrempf A. Fitness and aging in Cardiocondyla obscurior ant queens. Curr Opin Insect Sci. 2016;16:58–63.
Rodrigues MA, Flatt T. Endocrine uncoupling of the trade-off between reproduction and somatic maintenance in eusocial insects. Curr Opin Insect Sci. 2016;16:1–8.
Rueppell O, Aumer D, Moritz RFA. Ties between ageing plasticity and reproductive physiology in honey bees (Apis mellifera) reveal a positive relation between fecundity and longevity as consequence of advanced social evolution. Curr Opin Insect Sci. 2016;16:64–8.
Toth AL, Sumner S, Jeanne RL. Patterns of longevity across a sociality gradient in vespid wasps. Curr Opin Insect Sci. 2016;16:28–35.
Negroni MA, Jongepier E, Feldmeyer B, Kramer BH, Foitzik S. Life history evolution in social insects: a female perspective. Curr Opin Insect Sci. 2016;16:51–7.
Komiya Y, Habas R. Wnt signal transduction pathways. Organ. 2008;4:68–75.
Kitano J, Lema SC, Luckenbach JA, Mori S, Kawagishi Y, Kusakabe M, Swanson P, Peichel CL. Report adaptive divergence in the thyroid hormone signaling pathway in the stickleback radiation. Curr Biol. 2010;20:2124–30.
Eijkelenboom A, Burgering BMT. FOXOs: signalling integrators for homeostasis maintenance. Nat Rev Mol Cell Biol. 2013;14:83–97.
Gould SJ. Wonderful life: the burgess shale and the nature of history. New York: W.W. Norton & Company; 1989.
Acknowledgements
The authors would like to thank several anonymous reviewers for helpful comments on the content of this study.
Funding
This project was funded by a grant to Barbara Feldmeyer and Susanne Foitzik (DFG FE 1333/3–1 and FO 298/17–1) and support from the E. N. Huyck Preserve, Rensselearville NY. Collection permits were obtained at the Parks and Preserves.
Availability of data and materials
All data analysed during this study are included in various Additional files (see below) and the contig ortholog clusters are available from NCBIO BioProject GSE95604 (GSE95604_2017July_TemnoClusters.fas-gb.tar.gz).
Author information
Authors and Affiliations
Contributions
DE and AA assembled the transcriptomes and created the ortholog clusters. DE and BF conducted the selected genes analyses. BF drafted the manuscript, all authors read and edited the manuscript and approved it before submission.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Information on sample collection sites and year for each species. (DOCX 112 kb)
Additional file 2:
Summary of read counts and contig information per species. (DOCX 14 kb)
Additional file 3:
Codeml results on selected gene identified on branches leading to slavemakers and hosts, as well as each single species. Plus information on shared and private selected genes. (XLSX 135 kb)
Additional file 4:
Results of randomisation statistics. (DOCX 16 kb)
Additional file 5:
KEGG pathways assigned to positively selected genes per branch. (XLSX 21 kb)
Additional file 6:
Summary shared selected genes between slavemaker-host species pairs. (XLSX 39 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Feldmeyer, B., Elsner, D., Alleman, A. et al. Species-specific genes under selection characterize the co-evolution of slavemaker and host lifestyles. BMC Evol Biol 17, 237 (2017). https://doi.org/10.1186/s12862-017-1078-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12862-017-1078-9