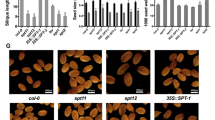
Abstract
Seed size is an important trait in determinant of rice seed quality and yield. In this study, we report a novel semi-dominant mutant S mall and r ound s eed 5 (Srs5) that encodes alpha-tubulin protein. Lemma cell length was reduced in Srs5 compared with that of the wild-type. Mutants defective in the G-protein alpha subunit (d1-1) and brassinosteroid receptor, BRI1 (d61-2) also exhibited short seed phenotypes, the former due to impaired cell numbers and the latter due to impaired cell length. Seeds of the double mutant of Srs5 and d61-2 were smaller than those of Srs5 or d61-2. Furthermore, SRS5 and BRI1 genes were highly expressed in Srs5 and d61-2 mutants. These data indicate that SRS5 independently regulates cell elongation of the brassinosteroid signal transduction pathway
Similar content being viewed by others
Background
Seed size and weight are important traits for rice yield (Song and Ashikari 2008, Takeda and Matsuoka 2008). Several quantitative trait loci (QTLs) affecting seed size have been identified, namely GW2 encoding a RING-type protein that functions as an E3 ubiquitin ligase (Song et al. 2007), qSW5 encoding a novel protein with no known domains (Shoumura et al. 2008), and GS3 encoding a membrane protein with various conserved domains (Fan et al. 2006, Takano-Kai et al. 2009). Loss of GW2 and qSW5 function leads to a wider seed phenotype, and loss of GS3 function leads to a longer seed phenotype, both resulting in increased yield.
Causal genes of the small (or short) seed mutants have also been identified, namely d1 (also named RGA1) encoding the heterotrimeric G protein alpha subunit (Ashikari et al. 1999, Fujisawa et al. 1999), d11 encoding a cytochrome P450 involved in brassinosteroid (BR) biosynthesis (Tanabe et al. 2005), d2 and brd2 encoding another type of cytochrome P450 involved in BR synthesis (Hong et al. 2003, Hong et al. 2005), d61 (also named OsBRI1) encoding the BR receptor (Yamamuro et al. 2000), srs1 encoding a novel protein that has no known functional domains (Abe et al. 2010), and finally, srs3 encoding a kinesin 13 protein (Kitagawa et al. 2010). During seed formation in rice, it was demonstrated that D1 regulates cell number (Izawa et al. 2010), and SRS1 and SRS3 regulate cell length (Abe et al. 2010, Kitagawa et al. 2010). From these observations, SRS1 and SRS3 seem to affect seed size through signaling pathways other than G-protein signal transduction.
Although several genes regulating seed size have been identified, their molecular network underlying seed formation remains unclear. Here we report molecular cloning of a novel small and round seed mutant in Srs5 (S mall and r ound s eed 5). The results clearly demonstrated that Srs5 encodes alpha-tubulin and regulates cell elongation in rice seed.
Results
Characterization of the Srs5 mutant
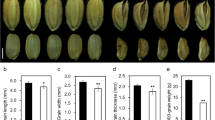
A mutant line, Kyudai No. 37, was identified by screening of small or short seed mutants from the rice collections of Togo Field, Nagoya University, and renamed S mall and r ound s eed 5, (Srs5). Srs5 shows shorter and rounder seeds, a shorter panicle and semi-dwarf plant phenotype, compared to WT (Figure 1A-C). Additionally, F1 plants derived from a cross between WT and Srs5 plants show intermediate seed length of parents seeds (Figure 1A and 1D). From these results, we presumed that the Srs5 mutation acts as semi-dominant gene. These phenotypes, short seed, short panicle, and dwarfism, are also exhibited by d1-1 and d61-2 mutants (Figure 1A-D). Comparison of internode elongation patterns among Srs5, d1-1, and d61-2 revealed that the internode elongation pattern of Srs5 differs from that of d1-1 and d61-2 (Figure 1E). Although d1-1 and d61-2 exhibit extremely stunted second, third, and fourth internodes, Srs5 shows equally shortened internodes (Figure 1E).
Srs5 mutant phenotypes. (A) Seed morphology of T65, Srs5, Srs5/SRS5, d1-1, and d61-2. Bar = 5 mm. (B) Panicle morphology of T65, Srs5, Srs5/SRS5, d1-1, and d61-2. Arrowheads indicate panicle neck nodes. Bar = 10 cm. (C) Gross morphology of T65, Srs5, Srs5/SRS5, d1-1, and d61-2. Bar = 20 cm. (D) Seed length of T65, Srs5, Srs5/SRS5, d1-1, and d61-2. Numbers on graphs indicate average seed length ± S.D. (E) Internode length relative to the total length of the culm. Schematic representation of the internode elongation pattern of T65, Srs5, d1-1, and d61-2. IN: internode.
To characterize short seed phenotype of Srs5 in detail, we compared the length of the inner epidermal cells of lemmas of Srs5, d1-1, and d61-2 using scanning electron microscopy (SEM). The cells of Srs5 were shorter than those of the WT (Figure 2A, B, and 2F), and similar to those of d61-2 (Figure 2D and 2F), but not those of d1-1 (Figure 2C and 2F). Additionally, we estimated cell numbers by dividing lemma length (Figure 2E) and by cell length (Figure 2F). Although d1-1 had a reduced number of inner epidermal cell of the lemma, the cell numbers of Srs5 and d61-2 were not significantly different from that of the WT (Figure 2G). From these observations, we concluded that the cause of the short seed phenotype of Srs5 is reduced cell length, as in d61-2.
Comparison of inner epidermal cell length of T65, Srs5, d1-1 , and d61-2. Inner epidermal cells of the lemma of WT (A), Srs5 (B), d1-1 (C), and d61-2 (D) observed by SEM. Bars = 100 μm. (E) Lemma length of the WT, Srs5, d1-1, and d61-2. (F) Inner epidermal cell length of WT, Srs5, d1-1, and d61-2. (G) Estimated cell numbers of WT, Srs5, d1-1, and d61-2. Numbers in (E-G) indicate averages ± S.D.
SRS5 gene encodes alpha-tubulin
To map the Srs5 locus on rice chromosomes, we performed linkage analysis using F2 plants derived from a cross between the Srs5 mutant (Oryza sativa. ssp. japonica) and Kasalath (Oryza sativa. ssp. indica). Since F2 seeds show a continuous variation in seed length, short seed phenotypic expression of seed size seems to be affected by difference in genetic background between the japonica and indica subspecies, in addition to Srs5 locus (Figure 3A). From 2000 F2 plants, we obtained 13 F2 plants that produced evident short seeds in F3 progeny, indicating homozygous of Srs5 mutant gene. Linkage analysis revealed that Srs5 was located in the 1.8 Mb between chr11-7240 and chr11-9030 on chromosome 11 (Figure 3B). To further fine mapping of the gene, it was difficult to perform linkage analysis using this population, because of small number of plants with homozygous of mutant allele (small seed). For further analysis, we produced an F2 population derived from a cross between Srs5 and a C hromosome S egment S ubstitution L ine (CSSL), SL233 that possessed the Kasalath chromosome segment of the short arm of chromosome 11 in a Koshihikari chromosome background. The phenotypes of F3 seeds obtained from these F2 plants were clearly distinguished mutant and wild types. From randomly selected 64 F2 plants, we obtained 24 WT plants (SRS5/SRS5), 36 heterozygote plants (Srs5/SRS5), and four mutant homozygote plants (Srs5/Srs5) (Figure 3C) with detecting the genotypes by using the PCR markers chr11-7240 and chr11-9030. The average seed lengths of SRS5/SRS5, Srs5/SRS5, and Srs5/Srs5 in the F2 population were 7.40 ± 0.15 mm, 6.67 ± 0.24 mm, and 5.84 ± 0.13 mm, respectively (Figure 3C). Since Srs5/SRS5 plants show seed lengths in-between those of SRS5/SRS5 and Srs5/Srs5 plants, the Srs5 mutant gene is considered to act as a semi-dominant manner (Figures 1A and 1D and 3C). The molecular markers chr11-7240 and chr11-9030 were used as selection markers for large-scale mapping (Figure 3B and 3D). We selected 664 from 5184 F2 recombinant plants that possessed chromosomal recombination within these two markers. We designed new molecular markers between chr11-7240 and chr11-9030, and 664 plants were used for high-resolution mapping (Figure 3D). The genotypes of F2 plant were confirmed by observation of the seed of F3 and F4 progeny. Finally, we obtained three recombinants within the 11 kb region including two predicted genes (RAP-DB http://rapdb.dna.affrc.go.jp/) (Figure 3D). Sequence analysis revealed only one single nucleotide polymorphism (SNP) in Os11g0247300 (Figure 3D). This mutation was resulted in an amino-acid substitution from Arg to Leu in residue 308 (Figure 3D).
Positional cloning of the Srs5 gene. (A) Phenotypic distribution of F3 seed length obtained from F2 plants derived from a cross between Srs5 and Kasalath. Arrowheads indicate the average for WT, Srs5, and Kasalath. (B) Srs5 was mapped on the short arm of chromosome 11. (C) Phenotypic distribution of F3 seed length obtained from F2 plants derived from a cross between Srs5 and SL233. Arrowheads indicate the average for Srs5/Srs5 (white), Srs5/SRS5 (grey), and SRS5/SRS5 (black). (D) Linkage analysis of Srs5.
Complementation test
To confirm the SNP as causal mutation of Srs5, we performed a complementation test. The WT SRS5 gene was transformed into the Srs5 mutant. The seeds of positive transgenic plants were significantly longer than the seeds of those containing empty vector (Figure 4A and 4C). The dwarf phenotype was also rescued (Figure 4B), but phenotype of transgenic plant was not completely same as wild type.
SRS5 expression
Accumulation of SRS5 mRNAs during the rice life cycle was investigated. SRS5 mRNA accumulated in the shoot apex, young panicles, and spikelets in both the WT and the Srs5 mutant (Figure 5A). In spikelets, accumulation of SRS5 mRNA was higher in the Srs5 mutant than in the WT (Figure 5A). Additionally, we compared the expression levels of SRS5 and BRI1 genes among Srs5, d1-1, and d61-2 mutants. Interestingly, we detected higher accumulation of SRS5 and BRI1 genes in both of Srs5 and d61-2 mutants (Figure 5B and 5C). In d1-1 mutant, the mRNA amounts of these genes were same level as WT (Figure 5B and 5C).
Expression of SRS5. (A) Relative amounts of SRS5 mRNA in various organs were analyzed by quantitative RT-PCR. Abbreviations: S, Shoot; R, Root; SA, Shoot apex; LB, Leaf blade; LS, Leaf sheath; LJ, Lamina joint; RA, Rachis; SP, Spikelet. (B) Relative amounts of SRS5 mRNA in spikelet of WT, Srs5, d1-1, and d61-2. (C) Relative amounts of BRI1 mRNA in spikelet of WT, Srs5, d1-1, and d61-2. Bars represent S.D.
Srs5 regulates cell elongation independently of BR signal transduction
Since Srs5 exhibits shorter cells, as does d61-2, we produced a double mutant by crossing to determine whether these two genes have epistasis. The double mutant showed shorter seed length (Figure 6A and 6C) and plant height than both parent mutants (Figure 6B). This result indicates that SRS5 and D61 regulate cell elongation independently during seed formation.
Epistatic test of Srs5 and d61-2. (A) Seed morphology of T65, Srs5, d61-2, and Srs5×d61-2. Bar = 5 mm. (B) Gross morphology of T65, Srs5, d61-2, and Srs5×d61-2. Bar = 20 cm. (C) Seed length of T65, Srs5, d61-2, and Srs5×d61-2. Numbers indicate averages ± S.D. (D) Genetic model of SRS5 function during seed elongation in rice.
Discussion
In linkage analysis, we could not clearly distinguish seed size in F2 seed derived from distant cross between Srs5 and Kasalath. This was likely to be caused by background difference between indica and japonica. To overcome this, we used a chromosome segment substitution line (CSSL), which is a plant series that possesses relatively large chromosome segments of donor parent chromosomes in the recurrent parental chromosome background (Yano and Sasaki 1997, Yano 2001, Ebitani et al. 2005, Ashikari and Matsuoka 2006, Fukuoka et al. 2010). CSSLs can be used to achieve high accuracy in phenotyping in F2 populations. In fact, we could make classification of two seed size, wild and mutant type, in the F2 population derived from a cross between Srs5 and a CSSL.
Genetic analysis of the F2 population derived from a cross between Srs5 and SL233 demonstrated that the Srs5 gene act as semi-dominant manner. This semi-dominant effect was also confirmed in complementation test. Although The Srs5 mutants carrying WT SRS5 gene showed longer seeds than that of the plants containing empty vector, the degree of recovery was not completely same as WT (Figure 4B). The reason that the rescue by WT SRS5 gene was partial may be due to compete between WT and mutation gene products or incomplete conformation of the tubulin complex.
In this study, we demonstrated that the SRS5 gene encodes alpha-tubulin, which has been reported to be the causal gene of the rice mutation T w i sted d warf 1(Tid1) (Sunohara 2009). The Tid1 mutation acts as a semi-dominant gene by affecting the interaction of alpha and beta tubulin. Since Srs5 was also a semi-dominant mutation, it was likely caused by incomplete conformation of the tubulin complex. Tid1 shows right helical growth, in addition to a semi-dominant dwarf phenotype. Additionally, Arabidopsis Lefty1 and Lefty2 mutations in genes orthologous to SRS5 also show semi-dominant and left helical growth (Thitamadee et al. 2002). These two mutants were gain-of-function alleles and exhibited similar twisted plant phenotypes. As the Srs5 mutant does not exhibit a twisted phenotype, different mutations in alpha-tubulin seem to lead to different phenotypes. In spikelets, higher accumulation of SRS5 mRNA was detected in the Srs5 mutant than in the WT (Figure 5A). This seems to compensate for the reduced function of alpha-tubulin protein. Higher expression of SRS5 was also detected in d61-2 but not in d1-1 (Figure 5B and 5C). Furthermore, BRI1 gene highly expresses in Srs5 and d61-2 but not in d1-1 (Figure 5B and 5C). These results suggest that the expression of SRS5 and BRI1 genes are compensated by sensing the cell elongation inhibition in the SRS5 and d61-2 mutants, although SRS5 and BRI1 genes regulate cell elongation independently (Figure 6D). Three other alpha-tubulin genes are present in the rice genome, and they share a high homology (Sunohara et al. 2009). In organs that exhibited no significant change in phenotype in the Srs5 mutant, these alpha-tubulins might work redundantly to maintain rice body planning.
Conclusions
Our study demonstrated that short seed mutants can be classified into two types: those with reduced cell numbers, e.g., d1, and those with reduced cell length, e.g., d61 (Figure 2A, C, and 2D). This facilitates classification of novel seed mutants. The short seed phenotype of Srs5 was demonstrated to be caused by reduced cell length, as in d61; however, the additive phenotype of the double mutant indicated that SRS5 and D61 regulate seed length via different mechanisms. To evaluate the mechanisms regulating seed length, observation of microtubule arrangement and analysis of double mutants among srs1, srs3, Srs5, and various BR mutants need to be performed.
Methods
Plant materials and growth conditions
Kyudai No. 37 was first identified at Kyushu University and maintained in Togo Field, Nagoya University. Its genetic background is unknown. A japonica cultivar, Taichung 65, was used as the WT plant. d1-1 was identified as spontaneous mutant 'Daikoku' and substituted its genetic background into Taichung 65 by backcrossing with Taichung 65 as a recurrent parent at Kyushu University. d61-2 was obtained by MNU treatment of Taichung 65. F2 and two parental lines were sown at the beginning of April. The seedlings of all plants were transplanted at the beginning of May into a paddy field at the Research Center for Bioresources Development in Fukui, Japan. They were then grown under natural conditions. Transgenic plants were grown in a closed greenhouse under natural sunlight. Room temperature was maintained at 30°C from 09:00 to 18:00 and 25°C from 18:00 to 09:00.
Linkage analysis of SRS5
For mapping, F2 plants derived from a cross between Srs5 (japonica) and Kasalath (indica) were used. Genomic DNA was extracted from fresh leaf tissues of 13 F2 plants that exhibited the small and round seed phenotype by the CTAB method. The genetic linkage between the Srs5 locus and molecular markers was determined using the sequence tagged site (STS) and cleaved amplified polymorphic sequence (CAPS) markers reported by the Rice Genome Program and microsatellite markers (McCouch et al. 2002). F2 plants derived from a cross between Srs5 and SL233 were used for fine mapping of Srs5 gene. Recombinant plants possessing a recombination between PCR markers chr11-7240 and chr11-9030 were screened from 5184 F2 plants. Other markers on chromosome 11 were designed by comparing the sequences of Srs5 and SL233. Information on the PCR markers used in this study is shown in Table 1. Phenotypes were determined using F3 seeds obtained from F2 plants and F4 seeds obtained from F3 plants.
Production of transgenic plants
The BAC clone containing the SRS5 gene was screened from the BAC library (constructed by the CUGI BAC/EST Resource Center) using four PCR primers, alpha-tub-5kb-up, alpha-tub-intron, alpha-tub-exon, and alpha-tub-5kb-down.
The BAC clone OSJNBa0014D2 was partially digested by Sau 3AI and cloned into the Bam HI site of binary vector pYLTAC7 (Liu et al. 1999) (provided by RIKEN BioResource Center). This clone contains the 7.26 kb upstream region from the transcriptional start site of the Srs5 gene and the 1.13 kb downstream region from the end of the 3'UTR region of the SRS5 gene. The binary vector was transformed into Agrobacterium tumefaciens strain EHA105 (Hood 1993) by electroporation, and Srs5 mutants were transformed as reported previously (Ashikari et al. 2005). Srs5 mutants containing empty vectors were used as controls.
RNA isolation and RT-PCR
Total RNA was extracted using the RNeasy Mini Kit (QIAGEN, Hilden, Germany). cDNAs were synthesized from total RNA using the SuperScript III system (Invitrogen, Carlsbad, CA, USA).
For quantification of SRS5 mRNA, real-time RT-PCR was carried out using SYBR Premix Ex Taq ™ II (TAKARA Bio, Inc., Tokyo, Japan). Two primers, RT-alpha-tub, were used to quantify Srs5 expression; a further two, RT-OsUbiquitin, were used to quantify OsUbiquitin1 expression (accession No. Os06g0681400). The Thermal Cycler Dice Real Time System (TAKARA Bio, Inc.) was used for quantification for real-time RT-PCR.
References
Abe Y, Mieda K, Ando T, Kono I, Yano M, Kitano H, Iwasaki Y: The SMALL AND ROUND SEED1 (SRS1/DEP2) gene is involved in the regulation of seed size in rice. Genes Genet Syst 2010, 85: 327–39. 10.1266/ggs.85.327
Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Nishimura A, Angeles ER, Qian Q, Kitano H, Matsuoka M: Cytokinin oxidase regulates rice grain production. Science 2005, 309: 741–5. 10.1126/science.1113373
Ashikari M, Matsuoka M: Identification, isolation and pyramiding of quantitative trait loci for rice breeding. Trends Plant Sci 2006, 11: 344–50. 10.1016/j.tplants.2006.05.008
Ashikari M, Wu J, Yano M, Sasaki T, Yoshimura A: Rice gibberellin-insensitive dwarf mutant gene Dwarf 1 encodes the alpha-subunit of GTP-binding protein. Proc Natl Acad Sci USA 1999, 96: 10284–9. 10.1073/pnas.96.18.10284
Ebitani T, Takeuchi Y, Nonoue Y, Yamamoto T, Takeuchi K, Yano M: Construction and evaluation of chromosome segment substitution lines carrying overlapping chromosome segments of indica rice cultivar Kasalath in a genetic background of japonica elite cultivar Koshihikari. Breed Sci 2005, 55: 65–73. 10.1270/jsbbs.55.65
Fukuoka S, Nonoue Y, Yano M: Germplasm enhancement by developing advanced plant materials from diverse rice accessions. Breed Sci 2010, 60: 509–517. 10.1270/jsbbs.60.509
Fujisawa Y, Kato T, Ohki S, Ishikawa A, Kitano H, Sasaki T, Asahi T, Iwasaki Y: Suppression of the heterotrimeric G protein causes abnormal morphology, including dwarfism, in rice. Proc Natl Acad Sci USA 1999, 96: 7575–80. 10.1073/pnas.96.13.7575
Hong Z, Ueguchi-Tanaka M, Fujioka S, Takatsuto S, Yoshida S, Hasegawa Y, Ashikari M, Kitano H, Matsuoka M: The rice brassinosteroid-deficient dwarf2 mutant, defective in the rice homolog of Arabidopsis DIMINUTO/DWARF1, is rescued by the endogenously accumulated alternative bioactive brassinosteroid, dolichosterone. Plant Cell 2005, 17: 2243–54. 10.1105/tpc.105.030973
Hong Z, Ueguchi-Tanaka M, Umemura K, Uozu S, Fujioka S, Takatsuto S, Yoshida S, Ashikari M, Kitano H, Matsuoka M: A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 2003, 15: 2900–10. 10.1105/tpc.014712
Hood EE, Gelvin SB, Melches LS, Hoekema A: New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res 1993, 2: 208–218. 10.1007/BF01977351
Izawa Y, Minami M, Ohki S, Iwasaki Y: Expression profile of the α subunit of the heterotrimeric G protein in rice. Plant Signal Behav 2010, 5: 845–7. 10.4161/psb.5.7.11824
Kitagawa K, Kurinami S, Oki K, Abe Y, Ando T, Kono I, Yano M, Kitano H, Iwasaki Y: A novel kinesin 13 protein regulating rice seed length. Plant Cell Physiol 2010, 51: 1315–29. 10.1093/pcp/pcq092
Liu YG, Shirano Y, Fukaki H, Yanai Y, Tasaka M, Tabata S, Shibata D: Complementation of plant mutants with large genomic DNA fragments by a transformation-competent artificial chromosome vector accelerates positional cloning. Proc Natl Acad Sci USA 1999, 96: 6535–40. 10.1073/pnas.96.11.6535
McCouch SR, Teytelman L, Xu Y, Lobos KB, Clare K, Walton M, Fu B, Maghirang R, Li Z, Xing Y, Zhang Q, Kono I, Yano M, Fjellstrom R, DeClerck G, Schneider D, Cartinhour S, Ware D, Stein L: Development and mapping of 2240 new SSR markers for rice ( Oryza sativa L.). DNA Res 2002, 9: 199–207. 10.1093/dnares/9.6.199
Sunohara H, Kawai T, Shimizu-Sato S, Sato Y, Sato K, Kitano H: A dominant mutation of TWISTED DWARF 1 encoding an alpha-tubulin protein causes severe dwarfism and right helical growth in rice. Genes Genet. Syst 2009, 84: 209–18.
Tanabe S, Ashikari M, Fujioka S, Takatsuto S, Yoshida S, Yano M, Yoshimura A, Kitano H, Matsuoka M, Fujisawa Y, Kato H, Iwasaki Y: A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell 2005, 17: 776–90. 10.1105/tpc.104.024950
Thitamadee S, Tuchihara K, Hashimoto T: Microtubule basis for left-handed helical growth in Arabidopsis . Nature 2002, 417: 193–6. 10.1038/417193a
Yamamuro C, Ihara Y, Wu X, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Kitano H, Matsuoka M: Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 2000, 12: 1591–606.
Yano M: Genetic and molecular dissection of naturally occurring variation. Curr Opin Plant Biol 2001, 4: 130–5. 10.1016/S1369-5266(00)00148-5
Yano M, Sasaki T: Genetic and molecular dissection of quantitative traits in rice. Plant Mol Biol 1997, 35: 145–53. 10.1023/A:1005764209331
Acknowledgements
This work was supported in part by the Funding Program for Next Generation World-Leading Researchers (NEXT Program) [GS-024] and the Ministry of Agriculture, Forestry and Fisheries of Japan [Genomics for Agricultural Innovation IPG-0002].
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
SS carried out molecular genetic studies, expression analysis, electron microscopic analysis, and transgenic analysis, and wrote manuscript. IK carried out molecular genetic studies. TA carried out molecular genetic studies. MY carried out molecular genetic studies and wrote manuscript. HK provided all plant materials KM designed research, carried out molecular genetic studies, expression analysis, electron microscopic analysis, and transgenic analysis, and wrote manuscript. YI designed research and wrote manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Segami, S., Kono, I., Ando, T. et al. Small and round seed 5 gene encodes alpha-tubulin regulating seed cell elongation in rice. Rice 5, 4 (2012). https://doi.org/10.1186/1939-8433-5-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1939-8433-5-4