Abstract
Background
The protein-protein interaction network (PIN) is an effective information tool for understanding the complex biological processes inside the cell and solving many biological problems such as signaling pathway identification and prediction of protein functions. Eriocheir sinensis is a highly-commercial aquaculture species with an unclear proteome background which hinders the construction and development of PIN for E. sinensis. However, in recent years, the development of next-generation deep-sequencing techniques makes it possible to get high throughput data of E. sinensis tanscriptome and subsequently obtain a systematic overview of the protein-protein interaction system.
Results
In this work we sequenced the transcriptional RNA of eyestalk, Y-organ and hepatopancreas in E. sinensis and generated a PIN of E. sinensis which included 3,223 proteins and 35,787 interactions. Each protein-protein interaction in the network was scored according to the homology and genetic relationship. The signaling sub-network, representing the signal transduction pathways in E. sinensis, was extracted from the global network, which depicted a global view of the signaling systems in E. sinensis. Seven basic signal transduction pathways were identified in E. sinensis. By investigating the evolution paths of the seven pathways, we found that these pathways got mature in different evolutionary stages. Moreover, the functions of unclassified proteins and unigenes in the PIN of E. sinensis were predicted. Specifically, the functions of 549 unclassified proteins related to 864 unclassified unigenes were assigned, which respectively covered 76% and 73% of all the unclassified proteins and unigenes in the network.
Conclusions
The PIN generated in this work is the first large-scale PIN of aquatic crustacean, thereby providing a paradigmatic blueprint of the aquatic crustacean interactome. Signaling sub-network extracted from the global PIN depicts the interaction of different signaling proteins and the evolutionary paths of the identified signal transduction pathways. Furthermore, the function assignment of unclassified proteins based on the PIN offers a new reference in protein function exploration. More importantly, the construction of the E. sinensis PIN provides necessary experience for the exploration of PINs in other aquatic crustacean species.
Similar content being viewed by others
Background
The development of high throughput techniques supplies a rich source of information for the Protein-protein Interaction Network (PIN) research. The interpretation of such information is a key to understand the complex world of biological processes inside the cell [1]. Knowledge of PINs helps researchers to solve many problems such as signaling pathways identification [2], recognition of functional modules [3] and prediction of protein functions [4]. Given the significant importance of PINs, proteome-wide interaction networks based on protein interactions has been constructed for many organisms [5–7]. The early study of PIN mostly focused on Saccharomyces cerevisiae. Schwikowski et al. performed a global analysis of published proteins interactions in S. cerevisiae and predicted the functions of 364 previously uncharacterized proteins [8]. Some interesting sub-networks were extracted from the PINs of S. cerevisiae and analyzed, for example, the spindle pole body related sub-network in Ito T’s work [9] and DNA damage response data set in Ho Y’s work [10]. Construction and analysis of PINs for other microorganisms has been subsequently performed, such as the PINs of Drosophila melanogaster [11], Helicobacter pylori [12] and Bacillus subtilis [13]. In the decades-long development of PIN, interest has shifted from microbial systems [14, 15] to mammalian [16] and more kinds of organisms [5]. However, to date, there is no large-scale PIN available for the study of aquatic crustacean. Although much effort has been made on the phenotype or physiological study of aquatic animals and crustaceans [17], an important ongoing problem is that the original inducement of all the phenotype and physiological features is the expression of genes and interaction of proteins. However, the expression and interaction of genes and proteins are still indistinct in most aquatic animals. As the protein interactions based on the gene expression has a significant role in the in-depth exploration of the biological process mechanism in cells, a PIN is necessary and important for the systematic study of aquatic crustaceans.
The Chinese mitten crab (Eriocheir sinensis) (Henri Milne Edwards, 1854) is one of the most important aquaculture species in China with high commercial value as a food source [18]. Many studies have been performed focusing on single or several genes [19], proteins [20] or a specific pathway [21] to accelerate the growth or improve the immune and signal transduction system of E. sinensis. However, the genome sequence of any E. sinensis species is still unavailable. Therefore, a whole map of the protein interactions in E. sinensis is still fragmentary and different signaling pathways implicated in growth and immune response also remain incomplete. Recently, Illumina RNA-seq, the next-generation deep-sequencing technique, provides new approaches to obtain a whole transcriptome sequencing [22, 23], which makes it possible to get huge amounts of knowledge on E. sinensis proteins and subsequently obtain a systematic overview of the protein-protein interaction system.
In this work we sequenced the transcriptional RNA sequences in the eyestalk, Y-organ and hepatopancreas of E. sinensis and presented a substantial resource of affinity-tagged proteins. A PIN of E. sinensis was generated based on the transcriptome sequencing. The network covers hundreds of previously-uncharacterized proteins, thus providing functional associations and biological context for the proteins that previously lacked annotation. The signaling sub-network was extracted from the global PIN and the evolution paths of known signaling pathways were examined, which represents a new global view of the signaling systems in E. sinensis. Functional assignment of the unclassified proteins and unigenes supplies significant guidance for the in vivo investigation of proteins/genes related to specific function. To our knowledge, the PIN of E. sinensis is the first large-scale aquatic crustacean protein interaction network, thereby providing a systems biology view of an aquatic crustacean proteome.
Results and discussion
Transcriptome sequencing of E. sinensis
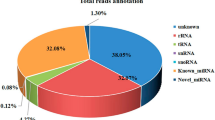
To obtain the E. sinensis transcriptome data, RNA from eyestalks, Y-organs and hepatopancreas mixed samples of E. sinensis were sequenced with the Illumina HiSeqTM2000. In total 2,358,728,280 nt clean nucleotides were found with Q20 and GC percentages of 96.68% and 45.08%, respectively. 26,208,092 clean reads were then obtained. From these clean reads, 157,168 contigs (mean length 236 nt) were assembled and then 58,582 unigenes (mean length 459 nt) were constructed from contigs with SOAP de novo, including 57,060 distinct singletons and 1,522 distinct clusters. The sequenced unigenes were subsequently aligned against the Nr database using BLASTn and BLASTx searching with E-value < 1*E-5. Finally 21,678 unigenes (37.00%) were matched. With Nr annotation, GO annotations of unigenes were obtained with the Blast2GO program. Among the total 58,582 unigenes of E. sinensis, 6,883 unigenes (11.75%) were annotated to the GO database with confident matches, including 4,680 assigned to the biological process category, 4241 assigned to the cellular component category and 5,684 assigned to the molecular function category. After the GO annotation of each unigene, WEGO software was used to obtain the GO functional classification for all unigenes in biological process category and to understand the distribution of gene functions from the macro level. In the biological process category, unigenes were divided into 26 different biological processes. Cellular process (3191; 68.2%) and metabolic process (2492; 53.2%) were most highly represented among them, other processed such as biological regulation (1392; 29.7%), developmental process (1094; 23.4%), localization (1166; 24.9%), multicellular organismal process (1170; 25%), regulation of biological process (1228; 26.2%) and response to stimulus (1057; 22.6%) were also included in biological process. The transcriptome sequencing and GO annotation results can be found in Additional file 1.
The protein information of model organisms
The protein sequence data of six model organisms (Drosophila melanogaster, Caenorhabditis elegans, Homo sapiens, Rattus norvegicus, Mus musculus, Saccharomyces cerevisiae) were downloaded from the Uniprot database. The number of protein sequences in each organism is shown in Table 1.
The protein interaction information of the above six model organisms was obtained from the PINA database. Usually, protein names were used in the protein interactions for presenting proteins, whereas for some interactions, the proteins in different model organisms were presented in different ways, which needed to be normalized for the convenience of the following analysis. Two special situations between protein IDs and protein names should be pretreated. Firstly, when there is no related protein name for a protein, the protein ID was used to mark the protein instead of a protein name. Secondly, sometimes the same protein is represented in different ways in different interactions. Some are marked with the protein name and the others are marked with the protein ID. In this situation, the protein name was used instead of the protein ID. For example, Vha36-2 is a protein in D. melanogaster. In the interaction pair: uniprotkb:Q7PLP5-uniprotkb:A1Z8V7, it is represented with the protein ID A1Z8V7 and in another interaction pair: uniprotkb:RpL15- uniprotkb:Vha36-2, it is represented with its protein name Vha36-2. We replaced A1Z8V7 with Vha36-2 in the first interaction to represent the protein. In this way, 44, 140, 679, 11, 10 and 40 protein names were pretreated in C. elegans, D. melanogaster, H. sapiens, M. musculus, R. norvegicus and S. cerevisiae, respectively. The number of proteins and protein-protein interactions of the six model organisms from PINA are shown in Table 2.
Construction of model-organism-based protein-protein interaction sub-network
In order to construct the PIN network of E. sinensis, the model-organism-based protein-protein interaction sub-networks were first constructed. The sub-networks were constructed based on the sequence alignment of model organisms and E. sinensis. According to the alignment result, the homologous sequence existing in each protein interaction of a model organism was marked. If two proteins in an interaction can both be matched with the unigenes in E. sinensis, then these two proteins and their interaction were considered as part of the model-organism-based sub-network. In this way, 6 different protein-protein interaction sub-networks were constructed based on the protein information of 6 model organisms respectively. The numbers of proteins, related unigenes and interactions in each model-organism-based protein-protein interaction sub-network are shown in Table 3. The unigenes and proteins in the sub-networks are not in one-to-one relationships. For example, protein yrt in D. melanogaste based sub-network is related to two unigenes (Unigene23137_A0A and Unigene37966_A0A) in E. sinensis.
Construction of PIN for E. sinensis
The PIN of E. sinensis was constructed according to the method described in the Methods section. The organism with a closer relationship to E. sinensis was integrated preferentially. D. melanogaster belongs to the Arthropoda-Insecta, which has the closest genetic relationship with E. sinensis (Arthropoda-Crustacea) compared with the other 5 organisms. C. elegans belongs to Protocoelomata-Nematomorpha, which is more primitive than Arthropoda-Crustacea, but has a closer genetic relationship with E. sinensis than the other 4 organisms. Therefore, D. melanogaster and C. elegans based sub-networks were used as the target and query networks respectively in the first turn integration. Subsequently the H. sapiens, R. norvegicus, M. musculus and S. cerevisiae based sub-network were aligned and integrated in order. One thing to mention is that R. norvegicus and M. musculus actually have similar genetic relationship with H. sapiens. However, the protein and interaction information of H. sapiens from Uniprot and PINA database is much more than that of R. norvegicus and M. musculus. Considering the attribution of the organisms to the PIN reconstruction, the H. sapiens based sub-network was integrated prior to R. norvegicus and M. musculus. Various numbers of protein interactions were added to the integrated network in each turn of integration and thus the PIN of E. sinensis was expanded based on the model organisms. Finally the PIN of E. sinensis was obtained after five-turn integrations of the above 6 sub-networks. The final E. sinensis PIN contained 3,223 proteins and 35,787 interaction pairs (Additional file 2). The scale of each intermediate integrated network after each turn of integration is shown in Table 4.
The PIN of E. sinensis is composed of a largest weakly connected component (LWCC) and several small components. As the LWCC contains most of the nodes and edges in the global PIN, the topological features of the LWCC were analyzed and compared with that of the model-organism-based sub-networks. The basic features of LWCCs are shown in Table 5. Actually, the features of LWCCs from different sub-networks are quite different from each other. For example, the number of nodes and edges in D. melanogaster based sub-network is much more than that in C. elegans based sub-network, and the diameter is smaller, which means the connectivity of the D. melanogaster based sub-network is stronger than that of the C. elegans based sub-network. The smaller path length further proved this conclusion. The S. cerevisiae based sub-network obviously has the strongest connectivity. Generally the topological parameter values of E. sinensis are between that of S. cerevisiae and the other five organisms, indicating that the PIN of E. sinensis assimilates the information in the six model organisms as well as eliminating the redundant information.
Score of protein-protein interaction pair
As the proteins (nodes) and interactions (edges) in the PIN came from different model organisms with various genetic relationships with E. sinensis, in order to represent the confidence of the interaction pairs, the score of each protein-protein interaction pair was evaluated according to the method described in the Methods section. The highest score is 33, reached by 5 interaction pairs. The highest score of edges and nodes are 14 and 10, respectively. Figure 1 shows the score distribution of the interaction pairs in the PIN of E. sinensis. 35.35% interaction pairs distribute in scores of 11–15. The interaction pairs with scores of 6–10 and 16–20 make up 27.83% and 21.54% of all the interaction pairs respectively. The score of each interaction pair is shown in Additional file 2.
The number of interaction pairs with scores above or equal to 30 is only 27 (Figure 2). These interactions exist in most of the model organisms, indicating their conservatism. For example, proteins Pros26.4, Mov34, Pros45, Tbp-1 and Rpn11 in these interactions are part of 26S protease. Mov34 is a regulation subunit and Rpn11 is a metal protease component of 26S protease. 26S protease is a huge protease complex widely found in many organisms [24]. Pros35, Mov34, CG5525, Tcp-1zeta, lwr and skpA are the hub proteins with their degree larger than 33, which exist in at least four model organisms and have important biological function in the metabolism by influencing many of their neighbor proteins.
Identification of signaling sub-network in E. sinensis
Signaling pathways are significant mediates of cell growth, development, immune and other life activities, which play a crucial role in almost all growth stages. To better understand the signaling systems of E. sinensis, the proteins with GO annotation “signaling” (GO:0023052) and their related interactions were extracted from the global PIN to generate the signaling sub-network. There are in total 572 proteins with GO:0023052 in E. sinensis, in which 67 are isolated and 505 interacted with other protein(s), the number of protein-protein interaction relationships is 2039 (Additional file 3). Seven known signal transduction pathways in the KEGG database were found in E. sinensis: Hippo, Jak-STAT mTOR, Wnt, MAPK, the Notch signal transduction pathway and the protein assembly process in endoplasmic reticulum. Some proteins are active in two or more signal pathways. Proteins in the above 7 pathways also interacted with other proteins in the PIN and finally the sub-network contains 313 proteins and 1,579 interaction pairs, including the 68 proteins in known signaling pathways and their neighboring proteins. The 68 proteins with 130 interaction pairs in known signaling pathways are shown in Figure 3A, in which 14 proteins and 1 interaction pair are isolated from the fully connected sub-network.
The Hippo signal transduction pathway is responsible for the growth inhibition of cells, which is a highly conservative pathway. It was first found in D. melanogaster and has been found in many mammals such as R. norvegicus and H. sapiens. The Hippo signal transduction pathway has significant function in organ size control, stem cell self-renewal, cancer inhibition and tissue homeostasis in response to multiple stimuli, including cell density and mechanotransduction [17, 25, 26]. Proteins wts, hpo and sav in this pathway are found to be responsible for cancer inhibition. The interaction of hpo and sav is able to phosphorylate and activate the complex composed of wts and Mats [27]. Two top cell skeleton signal proteins Mer and Ex can be reciprocally activated with kibra to further activate the Hippo pathway [28]. In addition, wts can directly phosphorylate, and thus inhibit the activating transcription factor yki. And yki is closely related with cell multiplication and apoptosis [25]. The Hippo related proteins wts, hpo, sav, Mats, Mer, yki and kibra were found in the signaling sub-network of E. sinensis (Figure 3B), indicating that the Hippo signal transduction pathway also exists in E. sinensis. The growth control and cell self-renewal of E. sinensis is probably dominated by the Hippo pathway.
The Jak-STAT signal transduction pathway is composed of the PTK related receptor, PTK JAK and transcription factor. It is simulated by cytokine and participates in many important biological processes such as cell multiplication, differentiation, apoptosis and immunoregulation [29]. The PIAS protein in this pathway can inhibit activation of the STAT protein by blocking the binding activity of the transcription factor and DNA. In addition, it is reported that PIAS can interact with more than 60 proteins, many of which are immune-system-related [30]. The STAT and PIAS proteins were found in the signal network of E. sinensis,coming from the H. sapiens based sub-network. The other proteins in the Jak-STAT pathway came from the D. melanogaster based sub-network, such as the suppressor of cytokine signaling (SOCS), which inhibits the phosphorylation of STAT by combining and blocking JAK or competing for the phosphotyrosine site on the cytokine receptor with STAT (Figure 3C). The multiplication, differentiation and apoptosis of E. sinensis are possibly controlled by the Jak-STAT pathway. The different source of proteins indicated that the integration process provided more information for the PIN of E. sinensis.
In addition, the mTOR, Wnt, MAPK, Notch and protein processing in endoplasmic reticulum were also found in the signaling sub-network of E. sinensis. The mTOR pathway is a central regulator for both cell proliferation and cell growth [31]. The Wnt pathway is involved in virtually every aspect of embryonic development and also controls homeostatic self-renewal in a number of adult tissues. Many studies report that mutation of the Wnt pathway is closely related to several hereditary diseases and cancers [32]. The Notch pathway is first found in D. melanogaster and participates in the regulation of cell multiplication, differentiation, and apoptosis, and acts as an important regulator of immune cells development [33]. The seven signaling transduction pathways found in E. sinensis represent the regulation of basic cell life activity about growth, development, reproduction and disease-resistance. The signaling sub-network of E. sinensis provides substantial information of the signal transduction pathways and unknown proteins which need to be further studied.
Evolution path of E. sinensis signaling network
The signaling network has been used to understand evolution in multicellular animals [34]. As the E. sinensis signaling sub-network was obtained from the integration of six model organisms and these organisms are located in different evolutionary stages, in order to promote understanding of the evolution of the signaling sub-network in E. sinensis, we examined the evolution path by comparing the E. sinensis signaling network with the six model organisms, and investigated the original organisms and preferred evolution paths of the E. sinensis signaling network. The six species were classified into three groups: primitive, bilaterian and vertebrate groups as described in Lei Li’s work [34]. The primitive group included S. cerevisiae. The bilaterian lineage was composed of D. melanogaster and C. elegans. All three vertebrate species were placed in the vertebrate group.
Based on this group partition, we identified each protein interaction in the E. sinensis signaling sub-network. For each protein interaction, the origin of two proteins and an interaction were defined separately using the principle in Lei Li’s work [34]. If a protein/interaction exists in a primitive organism, it is assigned to a primitive (or P) origin. If a protein/interaction exists in a bilaterian organism but not in a primitive organism, it is assigned to a bilaterian (or B) origin. If a protein/interaction exists only in vertebrate organism(s), it is assigned to a vertebrate (or V) origin. Finally, the origin of a complete protein interaction (including two proteins and an interaction) was assigned to the evolutionary stage in which the last component in the protein interaction appeared. The origin groups of seven known signaling pathways were examined (Figure 4). We found that different signaling pathways had various evolutionary paths. Protein processing in ER is relatively complete in the primitive stage with more than half of the interactions in this stage. Most interactions in the Hippo pathway exist in the bilateria and vertebrate stages, which is consistent with its first discovery in D. melanogaster [35]. Several interactions in the Hippo pathway originated from the primitive stage but no complete path was formed until it was found in D. melanogaster. The same situation also appears in the Jak-STAT, mTOR and Wnt pathway, indicating that these pathways may grow in the primitive stage and mature in the bilateria stages. MAPK and Notch have the latest evolution origin, with all interactions found in the biliteria and vertebrate stages.
Evolutionary origin of protein interactions in seven signaling pathways in study. Protein interactions in the seven signaling pathways in study were divided into “Evo-groups” according to the origins of the corresponding components. A blank in the right side represents that there are no protein originating in this evo-group. For each pathway, the proportion of interactions in each evolution stage to all the interactions in this pathway is shown in different shades of green. A darker green colour stands for a larger proportion.
Function assignment of unclassified proteins and unigenes
According to the GO biological process annotation of proteins in PIN, the functions of 2,496 proteins related to 4,981 unigenes were annotated, whereas the functions of the other 727 proteins related to 1,187 unigenes were still unknown, which makes up approximately 23% of all the proteins and 19% of all the unigenes. In order to investigate the functions of these unclassified proteins and unigenes, the method described in the Methods section was used to assign GO annotations. Finally, 549 unclassified proteins related to 864 unclassified unigenes were annotated (Figure 5), making up 76% and 73% of all the unknown proteins and unigenes (Additional file 4).
As the GO terms are organized in a treelike structure, we further analyzed the number distribution of newly-annotated proteins and unigenes in different GO depths (Figure 6). As GO depth increases, the annotation becomes more detailed. As shown in Figure 6, the fifth layer of the GO treelike structure contains the most assigned proteins and unigenes. As proteins or unigenes may have multiple functions, the GO annotation and proteins/unigenes are not in one-to-one relationship. 18 proteins were annotated as immune response-related proteins. They functioned in innate immune response (4 proteins), humoral immune response (2 proteins), mucosal immune response (5 proteins), regulation of innate immune response (6 proteins) and unclear immune response (7 proteins). Furthermore, 135 signaling-related proteins were found. They acted as receptors of signaling factor or the regulators of signaling pathways. For example, 16 proteins (Q8IQV6, Q7JXG9, Q8IR25, D3ZE26, Q9V3S7, Q9V3A8, Q7KMH9, Q7JPS2, Q9VVK8, Q9W1A7, Q7KN04, Q8SZY2, Q9VUP0, Q9VVU1, Q9VLK8, A1ZA45) were annotated as Hippo signaling cascade and 12 other proteins (Q8IQV6, Q7JXG9, Q8IR25, Q9V3S7, Q9V3A8, Q7KMH9, Q7JPS2, Q9VVK8, E1JII4, Q8SZY2, Q9VVU1, Q9VLK8) functioned as regulators of Hippo signaling cascade. Although the functions of the proteins/unigenes still need to be further validated, the assignment of functions provides important reference for identification of the targets in the in vivo experiment.
Conclusion
With the improvement in high-throughput sequencing technology, RNA sequencing and annotation are possible for further analysis and detection in the pursuit of certain biological goals. In present work we constructed a PIN of E. sinensis on the basis of transcriptomics sequencing and the proteome of six model organisms. The PIN defines a primary protein interaction landscape for E. sinensis cells that allows study of sub-networks with specific function. Seven known pathways were identified in the signaling sub-network extracted from the global PIN. With the analysis of evolution paths for these pathways, we found their differences in evolution origin. More proteins identified as neighbors of the proteins in seven identified pathways were prepared for further confirmation. Furthermore, the function assignment of unclassified proteins offers a new reference in protein function exploration. It is the first large-scale PIN of aquatic crustaceans, thereby providing necessary experience for the exploration of PIN for other aquatic crustacean species, as well as supplying a systems biological view of an aquatic crustacean interactome.
Methods
Obtaining of transcriptome data
Live E. sinensis (35–40 g in body weight) were purchased from the Tianjin Fisheries Institute and raised in fiberglass tanks. E. sinensis were cultured in freshwater at 18–20 degree centigrade (photoperiod L12:D12) for 7 days to acclimate to the laboratory conditions. Then three tissues including eyestalk, Y-organ and hepatopancreas were separated and collected. All samples were immediately frozen in liquid nitrogen and were stored at minus 80 degree centigrade until use. All experimental procedures were conducted in conformity with institutional guidelines for the care and use of laboratory animals in Tianjin Fisheries Institute and conformed to the National laboratory animal management regulations (Publication No. 2, 1988) approved by the National Science and Technology Commission.
Total RNA from E. sinensis tissue was sequenced with the Illumina high-throughput sequencing technology by Beijing Genomics Institution (BGI). The total RNA was extracted using the TRIzol method (Invitrogen) and then equal quantities of RNA from each tissue were pooled for transcriptome analysis. The samples for transcriptome analysis were prepared using Illumina’s kit and the generated library was sequenced using Illumina HiSeq™ 2000. Then the transcriptome de novo assembly was carried out with the short reads assembling program-Trinity [36] to generate unigenes. GO annotation of unigenes was obtained by the Blast2GO program [37] with an E-value cut-off at 1*E-5. WEGO software was used to obtain the GO functional classification for all unigenes in biological process category.
The protein sequences of model organisms
The protein sequence data of C. elegans, D. melanogaster, H. sapiens, M. musculus, R. norvegicus and S. cerevisiae was downloaded from the Uniprot database [38] (March 2012 version). The protein interactions of these model organisms were obtained from Protein-protein Interaction Network Analysis (PINA) [39, 40]. PINA integrates the protein interaction information of six public databases and supplies the complete, non-redundant protein interaction information of the above six model organisms. The March 2012 version was downloaded.
Gene ontology annotation
The Gene Ontology database [41] supplies a standardized representation of gene and gene product attributes across species and databases, including biological process, molecular function and cellular component. The gene_ontology.obo file was downloaded to obtain GO annotation from the Gene Ontology database. The GO annotation can be described as a directed acyclic graph according to the relations of GOs and a tree structure was drawn by programming. The GO numbers in each level of the tree were extracted.
Sequence alignment
The Basic Local Alignment Search Tool (BLAST) was downloaded from the NCBI ftp platform. The BLASTX program was used to align the nucleotide sequences (unigenes) in E. sinensis with the protein sequences of six model organisms to construct the model-organism-based protein-protein interaction sub-networks. The nucleotide sequence is first translated into protein sequences (one nucleotide sequence can be translated into six protein sequences) and then compared with the model organism one by one. The first aligned sequence with E value below 1*E-5 was considered as the homologous sequence.
Network integration
The construction of the PIN for E. sinensis is actually the integration of the 6 model-organism-based sub-networks. We developed an efficient computational procedure for integrating two PINs with reference to the global protein network alignment method in an attempt to obtain the integrated PIN [42]. The sub-networks were integrated one by one and the order was decided according to the genetic relationship of the model organisms with E. sinensis.
When two sub-networks were prepared to be combined, firstly, the homologies of the nodes in the two sub-networks were compared. Therefore, the two sub-networks were named as the target network and the query network. All the homologous proteins in the two networks were marked and the non-homologous proteins in the query network and the protein-protein relationships uniquely existing in the query network were added into the target network to form an integrated network. The detailed processes were as follows:
-
(1)
Proteins in the target and query networks were aligned with the BLASTP program, E value was set as 1*E-5.
-
(2)
The first matched protein in the target network to the query network was considered to be homologous. All the homologous proteins in the two networks were extracted.
-
(3)
The protein-protein interactions in the two networks were compared. When two proteins in an interaction pair in the query networks were both homologous to the target network (such as C-D in the target network and c-d in the query network in Figure 7), the protein names in the target network were used in the integrated network (such as C-D in Figure 7), and the new interaction pair in the query network was added if any (such as A-C in Figure 7); when only one protein in an interaction pair was homologous (such as D-E in the target network and d-g in the query network in Figure 7), then the protein name in the target network was used and the other protein in the interaction pair in the query was added (D-g in Figure 7); when no homologous proteins were found in an interaction pair in the query network, the protein names and this interaction in the query network were directly added into the integrated network.
-
(4)
The integrated network was considered as a new target network, and another model-organism-based sub-network was used as a new query network. Then steps (1) - (3) were repeated to generate a new integrated network. Such an iterative process was stopped until all the model-organism-based sub-networks were integrated. The final integrated network was the PIN of E. sinensis.
Topological features of networks
Diameter and average path length of network
In a directed network, the distance from node i to node j is the length of the shortest path between them. The diameter of a network is the length of the longest distance among all connected pairs of nodes in a graph. The average path length is the length of the distances averaged over all pairs of connected nodes in a graph [43].
Connected component
A strongly connected component (SCC) of a directed graph is a sub-graph where all nodes in the sub-graph are reachable by all other nodes in the sub-graph. Reachability between nodes is established by at least one directed path between the nodes. A weakly connected component (WCC) is a maximal group of nodes that are mutually reachable ignoring the edge directions [44].
The clustering coefficient
The clustering coefficient of a node v is defined as:
where k v is the number of nodes in the neighbourhood of vertex v,and e v is the number of edges existing between the neighbours of v. Suppose that a node v has k v neighbours, then at most k v (k v -1)/2 edges can exist between them (this occurs when every neighbour of v is connected to all the other neighbours of v). Let C v denote the fraction of these allowable edges that actually exist. The clustering coefficient of a network is defined as the average of C v over all v [45].
Degree and average degree
In graph theory, the degree of a graph is the number of edges incident to the nodes, with loops counted twice. The average degree is the degree averaged over all the nodes in a graph [44].
Index aggregation
The Index aggregation of a network is the ratio of the nodes in the largest WCC and the global network [44].
Score of protein-protein interaction pair
The protein interactions in E. sinensis PIN came from six model organisms. On one hand, proteins in different model organism were homologous to each other, as well as homologous to one or several unigenes in E. sinensis. On the other hand, organisms with close genetic relationship usually have similar protein interactions. Therefore, considering the homology of proteins and the genetic relationships of the model organisms with E. sinensis, the confidence score of the protein-protein interaction pairs in the PIN of E. sinensis was evaluated. Two factors were taken into consideration for scoring an interaction pair: point matching and edge matching. The point matching was actually the homology of the proteins in an interaction pair coming from different sub-networks. In the integration process, when a protein in the integrated network came from one sub-network, the score of this protein was 0. When the protein came from two homologous proteins in the target and query sub-networks, then the related unigene(s) of the two proteins were further checked. With the two proteins homologous to the same unigene(s) of E. sinensis, the score of their related protein in the integrated network was 2, otherwise it was 1. For example, in the first turn integration, protein CG6843 came from the integration of the homologous protein CG6843 in D. melanogaster and protein CIR-1 in C. elegans. Moreover, CG6843 and CIR-1 were both found to have high similarity with Unigene6670_A0A of E. sinensis by BLASTX. Therefore, the score of protein CG6843 in the integrated network was 2. Edge matching reflected the source of edge in the integrated network. The score of edges coming from the target network was higher than that from the query network because that the genetic relationship of the target organism was closer to E. sinensis than the query organism. Matched edge, which means the edge from both target and query network, was scored as 3. Mismatching edges from the target network and query network were scored as 2 and 1 respectively. Finally, the score of an interaction pair can be calculated as follows:
where S stands for the score of a protein-protein interaction pair; A and B are the score of two nodes (proteins) in an interaction pair respectively; R is the score of the edge; i stands for the number of integration times; N (N = 5) is the maximal number of interaction times. The maximum score of an interaction pair is 35 deduced by formula (1).
Function assignment of unclassified proteins
The functions of unknown proteins were annotated with the method mentioned in Alexei’s work [46]. The proteins were assigned to functional classes on the basis of their network of physical interactions as determined by minimizing the number of protein interactions among different functional categories [46]. Given one unknown function, we take the function that appears more often in the neighbor proteins of a known function as a prediction. Here a small change was made to Alexei’s method. In Alexei’s method, up to three of the most probable predicted functions were determined as final functions. While in the E. sinensis PIN, the number of function annotations for lots of known proteins is more than three. In avoid of missing important annotations, we use a parameter 25% instead of the top three rule. The detailed steps are as follows:
-
(1)
Identify the neighbor protein(s) interacting with the protein with unknown function (unclassified). The neighbor protein(s) with GO annotation were considered as classified protein(s);
-
(2)
Calculate the numbers of neighbor proteins with GO annotation and in the GO functional category;
-
(3)
If the number of neighbor proteins with a certain GO functional category make up more than 25% of the total number of neighbor proteins, then the GO annotation is assigned to the unclassified protein. If only one neighbor protein with GO annotations exists, all the GO annotations were assigned to the unclassified protein;
-
(4)
Taking into account the interactions among the above three steps, iterate (1)-(3) until no unclassified proteins can be further assigned.
References
Mosca R, Pons T, Ceol A, Valencia A, Aloy P: Towards a detailed atlas of protein-protein interactions. Curr Opin Struct Biol. 2013, 23: 929-940. 10.1016/j.sbi.2013.07.005.
Navlakha S, Gitter A, Bar-Joseph Z: A network-based approach for predicting missing pathway interactions. PLoS Comput Biol. 2012, 8: e1002640-10.1371/journal.pcbi.1002640.
Chen B, Fan W, Liu J, Wu FX: Identifying protein complexes and functional modules--from static PPI networks to dynamic PPI networks. Brief Bioinform. 2013, 15: 177-194.
Zeng E, Ding C, Narasimhan G, Holbrook SR: Estimating support for protein-protein interaction data with applications to function prediction. Comput Syst Bioinformatics Conf. 2008, 7: 73-84.
Guruharsha KG, Rual JF, Zhai B, Mintseris J, Vaidya P, Vaidya N, Beekman C, Wong C, Rhee DY, Cenaj O, McKillip E, Shah S, Stapleton M, Wan KH, Yu C, Parsa B, Carlson JW, Chen X, Kapadia B, VijayRaghavan K, Gygi SP, Celniker SE, Obar RA, Artavanis-Tsakonas S: A protein complex network of Drosophila melanogaster. Cell. 2011, 147: 690-703. 10.1016/j.cell.2011.08.047.
Kuhner S, van Noort V, Betts MJ, Leo-Macias A, Batisse C, Rode M, Yamada T, Maier T, Bader S, Beltran-Alvarez P, Castano-Diez D, Chen WH, Devos D, Guell M, Norambuena T, Racke I, Rybin V, Schmidt A, Yus E, Aebersold R, Herrmann R, Bottcher B, Frangakis AS, Russell RB, Serrano L, Bork P, Gavin AC: Proteome organization in a genome-reduced bacterium. Science. 2009, 326: 1235-1240. 10.1126/science.1176343.
Hu P, Janga SC, Babu M, Diaz-Mejia JJ, Butland G, Yang W, Pogoutse O, Guo X, Phanse S, Wong P, Chandran S, Christopoulos C, Nazarians-Armavil A, Nasseri NK, Musso G, Ali M, Nazemof N, Eroukova V, Golshani A, Paccanaro A, Greenblatt JF, Moreno-Hagelsieb G, Emili A: Global functional atlas of Escherichia coli encompassing previously uncharacterized proteins. PLoS Biol. 2009, 7: e96-10.1371/journal.pbio.1000096.
Schwikowski B, Uetz P, Fields S: A network of protein-protein interactions in yeast. Nat Biotechnol. 2000, 18: 1257-1261. 10.1038/82360.
Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y: A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 2001, 98: 4569-4574. 10.1073/pnas.061034498.
Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH: Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002, 415: 180-183. 10.1038/415180a.
Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, Pochart P, Machineni H, Welsh M, Kong Y, Zerhusen B, Malcolm R, Varrone Z, Collis A, Minto M, Burgess S, McDaniel L, Stimpson E, Spriggs F, Williams J, Neurath K, Ioime N, Agee M, Voss E, Furtak K, et al: A protein interaction map of Drosophila melanogaster. Science. 2003, 302: 1727-1736. 10.1126/science.1090289.
Kim KK, Kim HB: Protein interaction network related to Helicobacter pylori infection response. World J Gastroenterol. 2009, 15: 4518-4528. 10.3748/wjg.15.4518.
Marchadier E, Carballido-Lopez R, Brinster S, Fabret C, Mervelet P, Bessieres P, Noirot-Gros MF, Fromion V, Noirot P: An expanded protein-protein interaction network in Bacillus subtilis reveals a group of hubs: Exploration by an integrative approach. Proteomics. 2011, 11: 2981-2991. 10.1002/pmic.201000791.
Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM: A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000, 403: 623-627. 10.1038/35001009.
Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, Hao T, Rual JF, Dricot A, Vazquez A, Murray RR, Simon C, Tardivo L, Tam S, Svrzikapa N, Fan C, de Smet AS, Motyl A, Hudson ME, Park J, Xin X, Cusick ME, Moore T, Boone C, Snyder M, Roth FP, et al: High-quality binary protein interaction map of the yeast interactome network. Science. 2008, 322: 104-110. 10.1126/science.1158684.
Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, et al: Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005, 437: 1173-1178. 10.1038/nature04209.
Couzens AL, Knight JD, Kean MJ, Teo G, Weiss A, Dunham WH, Lin ZY, Bagshaw RD, Sicheri F, Pawson T, Wrana JL, Choi H, Gingras AC: Protein interaction network of the Mammalian hippo pathway reveals mechanisms of kinase-phosphatase interactions. Sci Signal. 2013, 6: rs15-10.1126/scisignal.2004712.
Zhang Y, Sun Y, Liu Y, Geng X, Wang X, Wang Y, Sun J, Yang W: Molt-inhibiting hormone from Chinese mitten crab (Eriocheir sinensis): Cloning, tissue expression and effects of recombinant peptide on ecdysteroid secretion of YOs. Gen Comp Endocrinol. 2011, 173: 467-474. 10.1016/j.ygcen.2011.07.010.
Yu AQ, Jin XK, Guo XN, Li S, Wu MH, Li WW, Wang Q: Two novel Toll genes (EsToll1 and EsToll2) from Eriocheir sinensis are differentially induced by lipopolysaccharide, peptidoglycan and zymosan. Fish Shellfish Immunol. 2013, 35: 1282-1292. 10.1016/j.fsi.2013.07.044.
Yanhua Wang YZ, Sun Y, Liu Y, Geng X, Sun J: cloing and molecular structure analysis of crustacean hyperglycemic hormone (Ers-CHH) in Eriocheir sinensis. J Fish China. 2013, 37: 987-993.
Li X, Cui Z, Liu Y, Song C, Shi G: Transcriptome analysis and discovery of genes involved in immune pathways from hepatopancreas of microbial challenged mitten crab Eriocheir sinensis. PLoS One. 2013, 8: e68233-10.1371/journal.pone.0068233.
Wang Z, Gerstein M, Snyder M: RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009, 10: 57-63. 10.1038/nrg2484.
Marguerat S, Bahler J: RNA-seq: from technology to biology. Cell Mol Life Sci. 2010, 67: 569-579. 10.1007/s00018-009-0180-6.
Peters JM, Franke WW, Kleinschmidt JA: Distinct 19 S and 20 S subcomplexes of the 26 S proteasome and their distribution in the nucleus and the cytoplasm. J Biol Chem. 1994, 269: 7709-7718.
Zhao B, Tumaneng K, Guan KL: The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol. 2011, 13: 877-883. 10.1038/ncb2303.
Halder G, Johnson RL: Hippo signaling: growth control and beyond. Development. 2011, 138: 9-22. 10.1242/dev.045500.
Lai ZC, Wei X, Shimizu T, Ramos E, Rohrbaugh M, Nikolaidis N, Ho LL, Li Y: Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 2005, 120: 675-685. 10.1016/j.cell.2004.12.036.
Baumgartner R, Poernbacher I, Buser N, Hafen E, Stocker H: The WW domain protein Kibra acts upstream of Hippo in Drosophila. Dev Cell. 2010, 18: 309-316. 10.1016/j.devcel.2009.12.013.
Krebs DL, Hilton DJ: SOCS proteins: negative regulators of cytokine signaling. Stem Cells. 2001, 19: 378-387. 10.1634/stemcells.19-5-378.
Shuai K: Regulation of cytokine signaling pathways by PIAS proteins. Cell Res. 2006, 16: 196-202. 10.1038/sj.cr.7310027.
Liu Y, Yan X, Zhou T: TBCK influences cell proliferation, cell size and mTOR signaling pathway. PLoS One. 2013, 8: e71349-10.1371/journal.pone.0071349.
Clevers H: Wnt/beta-catenin signaling in development and disease. Cell. 2006, 127: 469-480. 10.1016/j.cell.2006.10.018.
Radtke F, MacDonald HR, Tacchini-Cottier F: Regulation of innate and adaptive immunity by Notch. Nat Rev Immunol. 2013, 13: 427-437. 10.1038/nri3445.
Li L, Tibiche C, Fu C, Kaneko T, Moran MF, Schiller MR, Li SS, Wang E: The human phosphotyrosine signaling network: evolution and hotspots of hijacking in cancer. Genome Res. 2012, 22: 1222-1230. 10.1101/gr.128819.111.
Pan D: The hippo signaling pathway in development and cancer. Dev Cell. 2010, 19: 491-505. 10.1016/j.devcel.2010.09.011.
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A: Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011, 29: 644-652. 10.1038/nbt.1883.
Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M: Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005, 21: 3674-3676. 10.1093/bioinformatics/bti610.
Hinz U: From protein sequences to 3D-structures and beyond: the example of the UniProt knowledgebase. Cell Mol Life Sci. 2010, 67: 1049-1064. 10.1007/s00018-009-0229-6.
Wu J, Vallenius T, Ovaska K, Westermarck J, Makela TP, Hautaniemi S: Integrated network analysis platform for protein-protein interactions. Nat Methods. 2009, 6: 75-77. 10.1038/nmeth.1282.
Cowley MJ, Pinese M, Kassahn KS, Waddell N, Pearson JV, Grimmond SM, Biankin AV, Hautaniemi S, Wu J: PINA v2.0: mining interactome modules. Nucleic Acids Res. 2012, 40: D862-D865. 10.1093/nar/gkr967.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G: Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000, 25: 25-29. 10.1038/75556.
Kelley BP, Sharan R, Karp RM, Sittler T, Root DE, Stockwell BR, Ideker T: Conserved pathways within bacteria and yeast as revealed by global protein network alignment. Proc Natl Acad Sci U S A. 2003, 100: 11394-11399. 10.1073/pnas.1534710100.
J.A.Bondy USRM: Graph theory with applications. Macmillan Press Ltd. 1976
Diestel R: Graph theory. N Y. 2005
Watts DJ, Strogatz SH: Collective dynamics of ‘small-world’ networks. Nature. 1998, 393: 440-442. 10.1038/30918.
Vazquez A, Flammini A, Maritan A, Vespignani A: Global protein function prediction from protein-protein interaction networks. Nat Biotechnol. 2003, 21: 697-700. 10.1038/nbt825.
Acknowledgements
This work was supported by National High-Tech Research and Development Program of China (863 programs, 2012AA10A401 and 2012AA092205), Grants of the Major State Basic Research Development Program of China (973 programs, 2012CB114405), National Natural Science Foundation of China (21106095, 61100124), National Key Technology R&D Program (2011BAD13B07 and 2011BAD13B04), Foundation of Introducing Talents to Tianjin Normal University and “131” Innovative Talents cultivation of Tianjin.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
TH designed the experiment and wrote the manuscript. ZZ performed the experiments. JS provided the overall project guidance, critical review and gave advice on validation of manuscript. YZ and YL contributed in the transcriptome sequencing and GO annotation. BW gave advices on the manuscript revision. XZ executed the cultivation of E. sinensis and supplied the tissue sample for sequencing. All authors read and approved the manuscript.
Electronic supplementary material
12918_2014_1294_MOESM1_ESM.xls
Additional file 1:Transcriptome sequencing and GO annotation. Unigenes obtained from transcriptome sequencing and GO annotations. Nr annotations are also listed. GO annotations are shown in three categories: biological process, molecular function and cellular component. (XLS 10 MB)
12918_2014_1294_MOESM2_ESM.xls
Additional file 2:PIN of E. sinensis. Scores and species source of all the proteins and interactions in E. sinensis. (XLS 4 MB)
12918_2014_1294_MOESM3_ESM.xls
Additional file 3:Signaling sub-network of E. sinensis. Protein interaction pairs in signaling sub-network. Pathway information is listed in column C. Isolated nodes in the sub-network are marked in column D. (XLS 60 KB)
12918_2014_1294_MOESM4_ESM.xls
Additional file 4:Assigned proteins and unigenes. Assigned proteins and related unigenes with GO annotations and GO depth information. (XLS 125 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Hao, T., Zeng, Z., Wang, B. et al. The protein-protein interaction network of eyestalk, Y-organ and hepatopancreas in Chinese mitten crab Eriocheir sinensis . BMC Syst Biol 8, 39 (2014). https://doi.org/10.1186/1752-0509-8-39
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1752-0509-8-39