Abstract
Background
Gene mutation is an important mechanism of myeloid leukemogenesis. However, the number and combination of gene mutated in myeloid malignancies is still a matter of investigation.
Methods
We searched for mutations in the ASXL1, CBL, FLT3, IDH1, IDH2, JAK2, KRAS, NPM1, NRAS, RUNX1, TET2 and WT1 genes in 65 myelodysplastic syndromes (MDSs) and 64 acute myeloid leukemias (AMLs) without balanced translocation or complex karyotype.
Results
Mutations in ASXL1 and CBL were frequent in refractory anemia with excess of blasts. Mutations in TET2 occurred with similar frequency in MDSs and AMLs and associated equally with either ASXL1 or NPM1 mutations. Mutations of RUNX1 were mutually exclusive with TET2 and combined with ASXL1 but not with NPM1. Mutations in FLT3 (mutation and internal tandem duplication), IDH1, IDH2, NPM1 and WT1 occurred primarily in AMLs.
Conclusion
Only 14% MDSs but half AMLs had at least two mutations in the genes studied. Based on the observed combinations and exclusions we classified the 12 genes into four classes and propose a highly speculative model that at least a mutation in one of each class is necessary for developing AML with simple or normal karyotype.
Similar content being viewed by others
Background
Myeloid leukemogenesis is a complex process that transforms a regulated hematopoietic stem or progenitor cell into a proliferative cell unable to differentiate. Several genetic alterations such as translocations, gene mutations and deletions play a role in this process. Balanced translocations generating gene fusion play a major leukemogenic role in some classes of leukemias. However, they are rare in myelodysplastic syndromes (MDSs) and the majority of adult acute myeloid leukemias (AMLs) have a normal karyotype (NK-AMLs). MDSs are a heterogeneous group of clonal diseases characterized by bone marrow dysplasia, various degrees of cytopenia and a risk of progression to AML [1]. The alterations that lead to these pre-leukemic disorders are poorly defined [2]. Several genes have been identified recently as mutated in MDSs and AMLs, including ASXL1, located in chromosome arm 20q, CBL, located in 11q, and TET2, located in 4q [3–11]. These mutations are associated with biological and prognostic features [9, 11].
Similarly, gene mutations are likely to play a role in the development of NK-AMLs. Indeed, the identification of mutations in several genes such as NPM1 and FLT3 has revealed prognosis subgroups and modified the clinical management of these leukemias [12, 13]. A recent whole-genome sequencing study of an AML case has revealed mutations in IDH1, which encodes the enzyme isocitrate dehydrogenase [14]. AMLs occur de novo or after a chronic phase (hereafter called primary and secondary, respectively). Such chronic phase can be an MDS. We have recently shown that mutations in ASXL1, in contrast to NPM1 mutations, are found in MDSs and in secondary AMLs [15].
Mutation data are in line with the multi-hit model proposed by Gilliland suggesting that many gene mutations play a role in leukemogenesis [16]. Now that more and more gene mutations are reported it is important to determine when they occur in the various types of malignant hematopoietic diseases and how they combine in the development of leukemia. To begin to answer these questions we searched for mutations in twelve selected genes in a panel of 129 MDS and AML samples.
Methods
Patients and samples
All patients signed an informed consent and the study was approved by the institutional review board ("Commission d'Orientation Scientifique") of Institut Paoli-Calmettes.
The 129 samples were selected on the absence of known balanced translocation, and in the absence of complex karyotype for the AML series (as defined by more than three alterations). They included 65 cases of MDS (Additional file 1 Table S1), comprising, according to the WHO criteria [17], 5 refractory anemia (RA), 13 refractory anemia with ring sideroblasts (RARS) (including one with myelofibrosis), 7 refractory cytopenia with multilineage dysplasia (RCMD), 16 refractory anemia with excess of blasts type 1 (RAEB1), 19 refractory anemia with excess of blasts type 2 (RAEB2) and 5 MDS-unclassified (MDS-U) cases. Six cases were secondary to hematopoietic or non-hematopoietic diseases (na in IPSS column, Additional file 2 Table S1). The majority of MDS samples were collected at the time of diagnosis; some were in therapeutic abstention of a known MDS and some were under symptomatic treatment. Seventeen cases were IPPS low risk (0), 23 were int-1 (0.5-1), 12 were int-2 (1.5-2) and 7 were high risk (≥2.5).
We studied 64 AMLs (Additional file 2 Table S2) including 46 primary cases and 18 transformations of a previous myeloid disease (the same cohort of 64 AML patients, listed in Additional file 2 Table S2, had been analyzed for mutations in the NPM1, FLT3, CEBPA, NRAS, KRAS, JAK2, and ASXL1 genes and the results already reported in ref. 15). The panel comprised 47 cases of NK-AML and 17 cases with either trisomy 8 (n = 14), 9q deletion (HD-0632), trisomy 11 (HD-0304) or 20q deletion (HD-0381) as a sole karyotypic abnormality.
Array-comparative genomic hybridization (aCGH) had been performed on almost all samples and had allowed the detection of deletions and breaks [9, 10, 15, 18, 19].
DNA sequencing
DNA sequencing of exon-coding sequences of ASXL1, CBL, FLT3, IDH1, IDH2, JAK2, KRAS, NPM1, NRAS, RUNX1, TET2 and WT1 was done as follows. PCR amplifications of bone marrow cell DNA were done in a total volume of 25 μl PCR mix containing at least 5 ng template DNA, Taq buffer, 200 μmol of each deoxynucleotide triphosphate, 20 pmol of each primer and 1 unit of Hot Star Taq (Qiagen). PCR amplification conditions were as follows: 95°C 10 min; 95°C 30 sec, 55°C 30 sec, 72°C 30 sec to 1 min depending on PCR product length for 35 cycles; 72°C 10 min. PCR products were purified using Millipore plate MSNU030. One microliter of the purified PCR products was used for sequencing using the Big Dye terminator v1.1 kit (Applied Biosystems) including the forward or reverse primer. After G50 purification, sequences were loaded on an ABI 3130XL automat (Applied Biosystems). The sequence data files were analyzed using both SeqScape and Phred/Phrap/Consed softwares and all mutations were confirmed on an independent PCR product. The exons studied were as follows: ASXL1 exon 12, CBL exons 8 and 9, FLT3 exons 14, 15 and 20, IDH exon 4, JAK2 exon 14, RAS exons 1 and 2, NPM1 exon 12, RUNX1 exons 1 to 8, TET2 exons 3 to 11, WT1 exons 7 and 9. The primers for sequencing are listed in [Additional file 3].
Results
Mutations in MDSs
We had previously shown that ASXL1 frameshift and nonsense mutations occurred in exon 12 [10]. ASXL1 exon 12 frameshift mutations (10 times the same p.Gly646Trpfsx12) were observed in 12 out of the 65 MDS cases (18.5%) including 1 out of 5 RA (20%), 2 out of 16 RAEB1 (12.5%) and 9 out of 19 RAEB2 (47.4%) (Additional file 1 Table S1). We found 12 cases with TET2 mutation (18.5%) and 4 with RUNX1 mutation (6.2%). One patient (HD-0311) had two TET2 mutations. TET2 mutations were frequent in RAEB1 (7/16, 43.8%). Mutations in RUNX1 and TET2 were mutually exclusive but both could associate with ASXL1 mutations: two cases showed both an ASXL1 and a TET2 mutation and three cases both an ASXL1 and a RUNX1 mutation. One case of ASXL1 deletion (HD-0190) and one case of TET2 deletion (HD-0145) have been reported [9, 10]. One case (HD-0232) had a break in RUNX1 detected by aCGH (not shown).
We did not find any FLT3, NPM1 or WT1 mutation. One MDS-U had a JAK2 mutation and one RCMD case a KRAS mutation. Five cases, all RAEB2, were mutated in CBL. In one of these the mutation was homozygous. One subtitution occurred in the case with trisomy 11 (HD-0264), and showed a 2/3 ratio with the wild-type residue, suggesting that the mutated allele was duplicated (Figure 1a). This is in agreement with a potential gain-of-function of the mutated CBL [20]. We found 5 IDH mutations in the 65 cases (7.7%), including 2 mutations in IDH1 and 3 in IDH2.
Examples of mutations in leukemia target genes. a. sequence of CBL demonstrating c.1096G>A base substitution in the forward sequence at the position indicated by an arrow in HD-0264 patient with trisomy 11. The mutation leads to Glu366Lys change in the protein (not tolerated, SIFT). Sequence numbering is according to Genbank accessions numbers NM_005188 and NP_005179. The same sequence from another patient without mutation is shown below for comparison. b. Novel IDH2 mutation in MDS and AML patients. Sequence of IDH2, demonstrating c.419G>A base substitution in the reverse sequence at the position indicated by an arrow in blood cells of patient HD-0104 and its absence in the patient's buccal smear. The mutation leads to Arg140Gln (R140Q) change in the protein (not tolerated, SIFT). Sequence numbering is according to Genbank accessions numbers NM_002168.2 and NP_002159.2.
Mutations in AMLs
We found heterozygous nonsense or frameshift mutations of ASXL1 in 11 out of the 64 cases and one case (HD-0381) was known to have a deletion (Additional file 2 Table S2) [15]. As previously shown [15]NPM1 mutations were found in 28 cases, mostly primary cases. We found 9 TET2 mutations in 9 cases, and 10 RUNX1 mutations in 9 cases (HD-0790 had two RUNX1 mutations). In addition, HD-0198 had a break in RUNX1 [19]. These alterations were found in both primary and secondary AMLs. Mutations in RUNX1 and TET2, RUNX1 and NPM1, and ASXL1 and NPM1 were mutually exclusive, respectively. TET2 mutations could associate with either mutated NPM1 or ASXL1.
Nineteen cases (mostly primary) had FLT3 internal tandem duplication or point mutation. Three cases had a RAS mutation and one a JAK2 V617F mutation. The only CBL mutation was found in the case with a trisomy 11 (HD-0304, the corresponding transformed state of HD-0264 MDS). Three primary cases (6.5%) showed a WT1 mutation. Three cases were mutated in IDH1. Four cases showed the Arg172Lys mutation in IDH2. We also found 11 cases (17%) with the Arg140Gln substitution in IDH2 (Figure 1b).
Paired cases
For two patients both the chronic (HD-0173, HD-0264) and acute (HD-0790, HD-0304) phases of the disease were available (in bold in Tables). RARS HD-0173 had a mutated RUNX1 allele. The corresponding AML (HD-0790) had two RUNX1 mutations. RAEB2 HD-0264 had a mutation in ASXL1, CBL and TET2 genes. In the corresponding AML we did not identify any additional mutation in the studied genes.
Summary of results
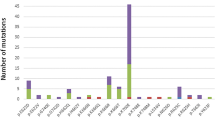
Additional file 4 Table S3 shows a summary of the results on mutated (cases with two mutations are counted as one) and deleted/broken cases. Mutations of ASXL1 were frequent in MDSs and secondary AMLs. Mutations of TET2 occurred with a similar frequency in MDSs and AMLs. ASXL1 and CBL mutations were associated with chronic phase, mainly RAEB2. Overall, advanced stage MDSs had more mutations. NPM1, FLT3 and WT1 mutations were associated with primary AMLs. Mutations of IDH1/2 were rare in chronic phase. Not surprisingly, the number of mutated genes per case was lower in MDSs than in AMLs: only 14% MDSs but more than half AMLs had at least two gene mutations.
Discussion
We searched for mutations in a series of selected genes. Mutations in some of these genes, such as ASXL1, CBL, IDH and TET2, have been identified only recently in myeloid diseases and have never been surveyed together to date. A number of issues need be discussed.
The frequencies of mutations we observed are close to what has been reported individually for each gene so far. This is true for example for IDH1, NPM1, TET2 and WT1 [1, 3–9, 14, 21, 22]. In MDS we found slightly more mutations of CBL than previously reported [11]. The IDH2 Arg140Gln was frequent in AMLs, rare in MDSs. The mutation was not present in the buccal smear DNA of a patient with AML (HD-0104), showing it was acquired. The Arg140Gln mutation in IDH2 has been reported in recent studies of myeloid diseases [23, 24]. A recent study has described ASXL1 as the most frequently mutated gene in advanced MDSs [25], which we confirmed here.
A repertoire of mutations
Based on the known functions of the proteins, on a previous model and classification [16], on where the mutations were present (MDSs and/or secondary AMLs and/or primary AMLs) and on how they combined (mutual exclusion or association), we tentatively grouped the genes in four classes. The first class includes RUNX1 and TET2. Mutations in these genes may cause clonal dominance of hematopoietic stem cells [3]. ASXL1 and NPM1 would constitute class II. Mutations in these genes may promote a pathway leading towards either primary or secondary AML [15]. Genes associated with signaling pathways and proliferation [16] (CBL, FLT3, JAK2, RAS) define class III. JAK2 mutation plays little role in MDSs and NK-AMLs. We have shown that FLT3 signaling is regulated by CBL [26]. However, the two alterations may not be equivalent; if in our study mutations in these genes were mutually exclusive and were both more frequent in AMLs than in MDSs, CBL but not FLT3 mutations were frequent in RAEB2 cases. This is in agreement with recent studies that showed alterations of CBL in chronic myeloid diseases [11, 27]. Finally, for three reasons we grouped IDH1, IDH2, and WT1 in a putative class IV. First, IDH and WT1 mutations were exclusive but could co-occur with mutations in genes from other classes. Second, they occurred primarily in AMLs and were rare in MDSs. Third, mutations of these genes could be associated with modifications of the HIF1 and oxygen-sensing pathways [28, 29]. Class IV mutations are rather associated with acute phase.
The existence of many cases with few or no mutations in the selected genes, especially in some classes of MDSs such as MDS-U, RA, RARS and RCMD but also in primary AMLs (half of the cases with no or one mutation), suggests that other genes remain to be studied or discovered. Indeed, our study did not include several target genes, such as CBFB [30], CDKN1A/B, CEBPA, ETV6, KIT, NF1, NFIA [31], P53, RB1 and UTX [32]; it did not include either a search for the recently discovered mutations of the EZH2 gene [33, 34]. Moreover, our aCGH analyses of the same series showed that some cases without mutation in the studied genes do have deletions of other cancer genes (for example CDKN1B, ETV6 and UTX [32] are deleted in HD-0205) [9, 10, 15]. Thus, the repertoire of altered genes in both MDSs and AMLs is likely to include many genes and it is probable that whole-genome sequencing studies will confirm this. Because of this, and although no case had four mutations, we propose that AML develops following - at least - four cooperating mutations, one from each class (Figure 2). This is highly speculative however; the identification of new target genes and the study of others will lead to a more precise picture. Also, because we did not study AMLs with balanced translocation or complex karyotype this model is proposed only for AML with simple or normal karyotype.
Hypothetical model summarizing frequent gene mutations in the development of two types of acute myeloid leukemia (AML). A hematopoietic stem cell (HSC) or a progenitor cell acquires a series of mutations beginning in class I genes (green). Different additional events (yellow, red and blue) induce clonal expansion and differentiation block and lead either towards secondary AML (sAML) via a patent preleukemic phase or AML without patent preleukemic phase. The order of events may vary.
Random or ordered accumulation?
An important question is whether the mutations occur in a necessary order or randomly until differentiation of the malignant hematopoietic clone is completely blocked. RARS HD-0173 (one RUNX1 mutation) and AML HD-0790 (two RUNX1 mutations) are instructive samples from the same patient. It is tempting to interpret this observation as a bi-allelic inactivation of the RUNX1 gene. RUNX1 mutations are frequent in chronic myelomonocytic leukemia [19] but rare in myeloproliferative neoplasms except at the acute phase [35]. In our series, mutations in TET2 were equally frequent in MDSs and AMLs. Taken together this suggests that inactivation of class I tumor suppressors (RUNX1, TET2) can intervene at any stage, i.e. early or at progression. In studies of AMLs secondary to myeloproliferative neoplasms (MPN) mutations of TET2 have indeed been shown to occur early or late [30, 36]. A recent study showed that ASXL1 mutations were present at the chronic stage and, in agreement with our findings, that ASXL1, JAK2 and TET2 have non overlapping contributions to myeloid transformation [36]. The order of occurrence of ASXL1, TET2 and RUNX1 mutations may vary in different diseases (MPNs and MDSs) and in different patients. However, the number of paired cases was much too low in our series to draw any firm conclusion. The study of more paired cases is necessary to definitely clear this issue. In contrast, some mutations such as IDH1 and IDH2 amino acid changes were detected at the AML rather than the MDS stage. Mutations in proliferation genes - except in CBL - were rare in MDS. Taken together this suggests that mutations in classes III and IV may occur late in leukemogenesis.
Myeloid malignancies and cancer genes
The study of myeloid malignancies may modify the classical view of cancer gene inactivation. IDH1 mutations do not result in a loss but in a change in activity [37]. This is in agreement with the fact that, in contrast to ASXL1, RUNX1 and TET2, we never observed break or deletion at IDH loci by aCGH analysis. WT1 is mutated but also overexpressed in AMLs. Mutations in CBL are associated with a gain-of-function [20]. NPM1 overexpression leads to increased cell growth [38]. We have not so far found two mutations (or mutation and deletion) in ASXL1 in the same sample. It is possible that ASXL1 mutations are also associated with a gain-of-function [39]. Only RUNX1 and TET2 may be inactivated in the two-hit fashion that corresponds to "classical" tumor suppressors.
Impact of mutations on prognosis
Due to relatively small numbers our purpose was not to address prognosis issues. Besides, it may be illusory to study prognostic impact of tumor suppressors in the absence of knowledge of yet unknown genes that may serve the same function. However, some features could be briefly noted. In contrast to TET2 mutations, which were relatively evenly distributed among IPSS classes, ASXL1, CBL and IDH mutations were associated with more aggressive MDSs. Lower risk MDSs had fewer mutations of these genes than higher risk MDSs. Half of the 12 MDS with int-2 IPSS showed ASXL1 mutations, and reciprocally half of the ASXL1 mutations were in int-2 cases. Four CBL mutations were found in int-2 cases. The 7 MDSs with high IPSS had an abnormal karyotype but few mutations (ASXL1 and RUNX1 in HD-0377, CBL in HD-0193); this is not surprising since karyotype status has an important weight on IPSS as one of its components. A long follow-up on disease evolution should tell if and how the mutational status impacts on MDS progression to AML and should be used to fine-tune IPSS.
Conclusion
We have reported here the first comprehensive study including recently discovered gene mutations in MDSs and AMLs. We have proposed a speculative model of cooperative leukemogenesis for AML with simple or normal karyotype, which needs to be confirmed by the study of more cases and completed by the discovery of more genes. As this it represents a step towards the necessary determination of the complete mutational status of myeloid malignant diseases.
References
Tefferi A, Vardiman JW: Myelodysplastic syndromes. N Eng J Med. 2009, 361: 1872-1885. 10.1056/NEJMra0902908.
Acquaviva C, Gelsi-Boyer V, Birnbaum D: Myelodysplastic syndromes: lost between two states?. Leukemia. 2010, 24: 1-5. 10.1038/leu.2009.157.
Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Massé A, Kosmider O, Le Couedic JP, Robert F, Alberdi A, Plo I, Dreyfus FJ, Marzac C, Casadevall N, Lacombe C, Romana SP, Dessen P, Soulier J, Viguié F, Fontenay M, Vainchenker W, Bernard OA: Mutation in TET2 in myeloid cancers. N Engl J Med. 2009, 360: 2289-2301. 10.1056/NEJMoa0810069.
Tefferi A, Lim KH, Abdel-Wahab O, Lasho TL, Patel J, Patnaik MM, Hanson CA, Pardanani A, Gilliland DG, Levine RL: Detection of mutant TET2 in myeloid malignancies other than myeloproliferative neoplasms: CMML, MDS, MDS/MPN and AML. Leukemia. 2009, 23: 1343-1345. 10.1038/leu.2009.59.
Jankowska AM, Szpurka H, Tiu RV, Makishima H, Afable M, Huh J, O'Keefe CL, Ganetzky R, McDevitt MA, Maciejewski JP: Loss of heterozygosity 4q24 and TET2 mutations associated with myelodysplastic/myeloproliferative neoplasms. Blood. 2009, 113: 6403-6410. 10.1182/blood-2009-02-205690.
Abdel-Wahab O, Mullally A, Hedvat C, Garcia-Manero G, Patel J, Wadleigh M, Malinge S, Yao JJ, Kilpivaara O, Bhat R, Huberman K, Thomas S, Dolgalev I, Heguy A, Paietta E, Le Beau MM, Beran M, Tallman MS, Ebert BL, Kantarjian HM, Stone RM, Gilliland DG, Crispino JD, Levine RL: Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009, 114: 144-147. 10.1182/blood-2009-03-210039.
Langemeijer SM, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M, Stevens-Linders E, van Hoogen P, Geurts van Kessel A, Raymakers RAP, Kamping EJ, Verhoef GE, Verburgh E, Hagemeijer A, Vandenberghe P, de Witte T, van der Reijden BA, Janssen JH: Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet. 2009, 41: 838-842. 10.1038/ng.391.
Mohamedali AM, Smith AE, Gaken J, Lea NC, Mian SA, Westwood NB, Strupp C, Gattermann N, Germing U, Mufti GJ: Novel TET2 mutations associated with UPD4q24 in myelodysplastic syndromes. J Clin Oncol. 2009, 27: 4002-4006. 10.1200/JCO.2009.22.6985.
Kosmider O, Gelsi-Boyer V, Cheok M, Grabar S, Della-Valle V, Picard F, Viguié F, Quesnel B, Beyne-Rauzy O, Solary E, Vey N, Hunault-Berger M, Fenaux P, Mansat-De Mas V, Delabesse E, Guardiola P, Lacombe C, Vainchenker W, Preudhomme C, Dreyfus F, Bernard OA, Birnbaum D, Fontenay M: TET2 mutation is an independent favorable prognostic factor in myelodysplastic syndromes (MDS). Blood. 2009, 114: 3285-3291. 10.1182/blood-2009-04-215814.
Gelsi-Boyer V, Trouplin V, Adélaïde J, Bonansea J, Cervera N, Carbuccia N, Lagarde A, Prébet T, Nezri M, Sainty D, Olschwang S, Xerri L, Chaffanet M, Mozziconacci MJ, Vey N, Birnbaum D: Mutations of polycomb-associated gene ASXL1 in myelodysplastic syndromes and chronic myelomonocytic leukaemia. Br J Haematol. 2009, 145: 788-800. 10.1111/j.1365-2141.2009.07697.x.
Makishima H, Cazzolli H, Szpurka H, Dunbar A, Tiu R, Huh J, Muramatsu H, O'Keefe C, Hsi E, Paquette RL, Kojima S, List AF, Sekeres MA, McDevitt MA, Maciejewski JP: Mutations of E3 ligase Cbl family members constitute a novel common pathogenic lesion in myeloid malignancies. J Clin Oncol. 2009, 27: 6109-6116. 10.1200/JCO.2009.23.7503.
Falini B, Bolli N, Liso A, Martelli MP, Mannucci R, Pileri S, Nicoletti N: Altered nucleophosmin transport in acute myeloid leukaemia with mutated NPM1: molecular basis and clinical implications. Leukemia. 2009, 23: 1731-1743. 10.1038/leu.2009.124.
Gaidzik V, Döhner K: Prognostic implications of gene mutations in acute myeloid leukemia with normal karyotype. Semin Oncol. 2008, 35: 346-355. 10.1053/j.seminoncol.2008.04.005.
Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, Fulton LA, Locke DP, Magrini VJ, Abbott RM, Vickery TL, Reed JS, Robinson JS, Wylie T, Smith SM, Carmichael L, Elred JM, Harris CC, Walker J, Peck JB, Du F, Dukes AF, Sanderson GE, Brummett AM, Clark E, McMichael JF, Meyer RJ, Schindler JK, Pohl CS, Wallis JW, Shi X, Lin L, Schmidt H, Tang Y, Haipek C, Wiechert ME, Ivy JV, Kalicki J, Elliott G, Ries RE, Payton JE, Westervelt P, Tomasson MH, Watson MA, Baty J, Heath S, Shannon WD, Nagarajan R, Link DC, Walter MJ, Graubert TA, DiPersio JF, Wilson RK, Ley TJ: Recurrent mutations found by sequencing an acute myeloid leukemia genome. N Eng J Med. 2009, 361: 1058-1066. 10.1056/NEJMoa0903840.
Carbuccia N, Trouplin V, Gelsi-Boyer V, Murati A, Rocquain J, Adélaïde J, Olschwang S, Xerri L, Vey N, Chaffanet M, Birnbaum D, Mozziconacci MJ: Mutual exclusion of ASXL1 and NPM1 mutations in a series of acute myeloid leukemias. Leukemia. 2010, 24: 469-473. 10.1038/leu.2009.218.
Gilliland DG: Molecular genetics of human leukemias: new insights into therapy. Semin Hematol. 2002, 39: 6-11. 10.1053/shem.2002.36921.
Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, Harris NL, Le Beau MM, Hellström-Lindberg E, Tefferi A, Bloomfield CD: The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009, 114: 937-951. 10.1182/blood-2009-03-209262.
Murati A, Gervais C, Carbuccia N, Finetti P, Cervera N, Adélaïde J, Struski S, Lippert E, Mugneret F, Tigaud I, Penther D, Bastard C, Poppe B, Speleman F, Baranger L, Luquet I, Cornillet-Lefebvre P, Nadal N, Nguyen-Khac F, Pérot C, Olschwang S, Bertucci F, Chaffanet M, Lessard M, Mozziconacci MJ, Birnbaum D: Genomic profiling of acute myelomonocytic leukemia: alteration of the MYB locusin MYST3-linked cases. Leukemia. 2009, 23: 85-94. 10.1038/leu.2008.257.
Gelsi-Boyer V, Trouplin V, Adélaïde J, Aceto N, Remy V, Pinson S, Houdayer C, Arnoulet C, Sainty D, Bentires-Alj M, Olschwang S, Vey N, Mozziconacci MJ, Birnbaum D, Chaffanet M: Genome profiling of chronic myelomonocytic leukemia: frequent alterations of RAS and RUNX1 genes. BMC Cancer. 2008, 8: 299-314. 10.1186/1471-2407-8-299.
Sanada M, Suzuki T, Shih LY, Otsu M, Kato M, Yamasaki S, Tamura A, Honda H, Sakata-Yanagimoto M, Kumano K, Oda H, Yamagata T, Takita J, Gotoh N, Nakazaki K, Kawamata N, Onodera M, Nobuyoshi M, Hayashi Y, Harada H, Kurokawa M, Chiba S, Mori H, Ozawa K, Omine M, Hirai H, Nakauchi H, Koeffler HP, Ogawa S: Gain-of-function of mutated CBL tumour suppressor in myeloid neoplasms. Nature. 2009, 460: 904-908. 10.1038/nature08240.
Renneville A, Boissel N, Zurawski V, Llopis L, Biggio V, Nibourel O, Philippe N, Thomas X, Dombret H, Preudhomme C: Wilms tumor gene mutations are associated with a higher risk of recurrence in young adults with acute myeloid leukemia: a study from the Acute Leukemia French Association. Cancer. 2009, 115: 3719-3727. 10.1002/cncr.24442.
Virappane P, Gale R, Hills R, Kakkas I, Summers K, Stevens J, Allen C, Green C, Quentmeier H, Drexler H, Burnett A, Linch D, Bonnet D, Lister TA, Fitzgibbon J: Mutation of the Wilm's tumor 1 gene is a poor prognostic factor associated with chemotherapy resistance in normal karyotype acute myeloid leukaemia: the United Kingdom Medical Research Council Adult Leukaemia Working Party. J Clin Oncol. 2008, 26: 5429-5435. 10.1200/JCO.2008.16.0333.
Green A, Beer P: Somatic mutations of IDH1 and IDH2 in the leukemic transformation of myeloproliferative neoplasms. New Engl J Med. 2010, 362: 369-370. 10.1056/NEJMc0910063.
Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett B, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, Rabinowitz JD, Carroll M, Su S, Sharp KA, Levine RL, Thompson CB: The common features of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting α-ketoglutarate to 2-hydoxyglutarate. Cancer Cell. 2010, 17: 1-10. 10.1016/j.ccr.2010.01.020.
Boultwood J, Perry J, Pellagatti A, Fernandez-Mercado M, Fernandez-Santamaria C, Calasanz MJ, Larrayoz MJ, Garcia-Delgado M, Giagounidis A, Malcovati L, Della Porta MG, Jädersten M, Killick S, Hellström-Lindbergh E, Cazzola M, Wainscoat JS: Frequent mutations of the polycomb-associated gene ASXL1 in the myelodysplastic syndromes and in acute myeloid leukemia. Leukemia. 2010, 24: 1062-1065. 10.1038/leu.2010.20.
Lavagna-Sévenier C, Marchetto S, Birnbaum D, Rosnet O: FLT3 signaling in hematopoietic cells involves CBL, SHC and an unknown P115 as prominent tyrosine-phosphorylated substrates. Leukemia. 1998, 12: 301-310. 10.1038/sj.leu.2400921.
Dunbar AJ, Gondek LP, O'Keefe CL, Makishima H, Rataul MS, Szpurka H, Sekeres MA, Wang XF, McDevitt MA, Maciejewski JP: 250K single nucleotide polymorphism array karyotyping identifies acquired uniparental disomy and homozygous mutations, including novel missense substitutions of c-Cbl, in myeloid malignancies. Cancer Res. 2008, 68: 10340-10357. 10.1158/0008-5472.CAN-08-2754.
Pollard PJ, Ratcliffe PJ: Puzzling patterns of predisposition. Science. 2009, 324: 192-194. 10.1126/science.1173362.
Wagner KD, Wagner N, Wellman S, Schley G, Bondke A, Theres H, Scholz H: Oxygen-regulated expression of the Wilm's tumor suppressor Wt1 involves hypoxia-inducible factor-1 (HIF-1). FASEB J. 2003, 17: 1364-1366.
Couronné L, Lippert E, Andrieux J, Kosmider O, Radford-Weiss I, Penther D, et al: Analyses of TET2 mutations in post-myeloproliferative neoplasm acute myeloid leukemias. Leukemia. 2010, 24: 201-203. 10.1038/leu.2009.169.
Bernard F, Gelsi-Boyer V, Murati A, Giraudier S, Trouplin V, Adélaïde J, Rey J, Olschwang S, Vainchenker W, Chaffanet M, Vey N, Mozziconacci MJ, Birnbaum D: Alterations of NFIA in chronic malignant myeloid diseases. Leukemia. 2009, 23: 583-585. 10.1038/leu.2008.228.
van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, Edkins S, Hardy C, O'Meara S, Teague J, Butler A, Hinton J, Latimer C, Andrews J, Barthorpe S, Beare D, Buck G, Campbell PJ, Cole J, Forbes S, Jia M, Jones D, Kok CY, Leroy C, Lin ML, McBride DJ, Maddison M, Maquire S, McLay K, Menzies A, Mironenko T, Mulderrig L, Mudie L, Pleasance E, Shepherd R, Smith R, Stebbings L, Stephens P, Tang G, Tarpey PS, Turner R, Turrell K, Varian J, West S, Widaa S, Wray P, Collins VP, Ichimura K, Law S, Wong J, Yuen ST, Leung SY, Tonon G, DePinho RA, Tai YT, Anderson KC, Kahnoski RJ, Massie A, Khoo SK, Teh BT, Stratton MR, Futreal PA: Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009, 41: 521-523. 10.1038/ng.349.
Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, Waghorn K, Zoi K, Ross FM, Reiter A, Hochhaus A, Drexler HG, Duncombe A, Cervantes F, Oscier D, Boultwood J, Grand FH, Cross NCP: Inactivating mutations of the histone methyltransferase gene EZH2in myeloid disorders. Nat Genet. 2010,
Nikoloski G, Langemeijer SMC, Kuiper RP, Knops R, Massop M, Tönnissen ERLTM, van der Heijden A, Scheele TN, Vandenberghe P, de Witte T, van der Reijden BA, Jansen JH: Somatic mutations of the histone methyltransferase gene EZH2in myelodysplastic syndromes. Nat Genet. 2010,
Ding Y, Harada Y, Imagawa J, Kimura A, Harada H: AML1/RUNX1 point mutation possibly promotes leukemic transformation in myeloproliferative neoplasms. Blood. 2009, 114: 5201-5205. 10.1182/blood-2009-06-223982.
Abdel-Wahab O, Mansouri T, Patel J, Harris K, Yao J, Hedvat C, Hedvat C, Heguy A, Bueso-Ramos C, Kantarjian H, Levine RL, Verstovsek S: Genetic analysis of transforming events that convert chronic myeloproliferative neoplasms to leukemias. Cancer Res. 2010, 70: 447-452. 10.1158/0008-5472.CAN-09-3783.
Dang L, White DW, Gross S, Bennett BD, Bittinger MAA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Vander Heiden MG, Su SM: Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009, 462: 739-744. 10.1038/nature08617.
Grisendi S, Mecucci C, Falini B, Pandolfi PP: Nucleophosmin and cancer. Nat Rev Cancer. 2006, 494: 493-505. 10.1038/nrc1885.
Fisher CL, Pineault N, Brookes C, Helgason CD, Ohta H, Bodner C, Hess JL, Humphries RK, Brock HW: Loss-of-function Additional sex coombs-like 1 mutations disrupt hematopoiesis but do not cause severe myelodysplasia or leukemia. Blood. 2010, 115: 38-46. 10.1182/blood-2009-07-230698.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2407/10/401/prepub
Acknowledgements
We are very grateful to the patients who participated to the study. This work was supported by Inserm, Institut Paoli-Calmettes and grants from the Fondation de France (DB, Comité Leucémie 2007-2009) and Association pour la Recherche contre le Cancer (n° 4929; n° 4992).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
JR, NC, VT, SR and SO obtained and analyzed gene sequencing data. AM, MN, ZT, VGB; MJM and NV provided samples and bioclinical data. NV, DB, VGB and MJM designed and supervised the study, analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Julien Rocquain, Nadine Carbuccia, Virginie Trouplin contributed equally to this work.
Electronic supplementary material
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Rocquain, J., Carbuccia, N., Trouplin, V. et al. Combined mutations of ASXL1, CBL, FLT3, IDH1, IDH2, JAK2, KRAS, NPM1, NRAS, RUNX1, TET2 and WT1 genes in myelodysplastic syndromes and acute myeloid leukemias. BMC Cancer 10, 401 (2010). https://doi.org/10.1186/1471-2407-10-401
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2407-10-401