Abstract
Background
Exome sequencing has become more and more affordable and the technique has emerged as an important diagnostic tool for monogenic disorders at early stages of investigations, in particular when clinical information is limited or unspecific as well as in cases of genetic heterogeneity.
Methods
We identified a consanguineous Pakistani family segregating an autosomal recessive phenotype characterized by muscular hypertrophy, mild mental retardation and skeletal abnormalities. The available clinical information was incomplete and we applied whole exome sequencing in an affected family member for the identification of candidate gene variants.
Results
Exome sequencing identified a previously unreported homozygous mutation in the acceptor splice site of intron 5 in the BSCL2 gene (c.574-2A > G). Expression analysis revealed that the mutation was associated with skipping of exon 6. BSCL2 mutations are associated with Berardinelli-Seip congenital lipodystrophy and a clinical re-evaluation of affected individuals confirmed the diagnosis.
Conclusions
Exome sequencing is a powerful technique for the identification of candidate gene variants in Mendelian traits. We applied this technique on a single individual affected by a likely autosomal recessive disorder without access to complete clinical details. A homozygous and truncating mutation was identified in the BSCL2 gene suggesting congenital generalized lipodystrophy. Incomplete phenotypic delineations are frequent limiting factors in search for a diagnosis and may lead to inappropriate care and follow-up. Our study exemplifies exome sequencing as a powerful diagnostic tool in Mendelian disorders that may complement missing clinical information and accelerate clinical diagnosis.
Similar content being viewed by others
Background
Mutation detection in genetic diseases is important for a definite diagnosis for genetic counseling, future prenatal diagnosis and therapy. Next generation sequencing technologies are increasingly being used as a diagnostic tool for genetic disorders and have been shown particularly successful in genetically heterogeneous phenotypes, or when ascertainment of clinical features and symptoms are unattainable [1–10]. Furthermore, exome sequencing is in many cases a cost-effective diagnostic method of patients with Mendelian traits compared to the interrogation of candidate genes one by one.
We report herein on a family segregating short and muscular stature, mild mental retardation and acromegalic appearance as an autosomal recessive trait. The clinical information was otherwise sparse. Using whole exome sequencing (WES), we identified a homozygous mutation in the acceptor splice site of intron 5 of the BSCL2 gene previously associated to congenital generalized lipodystrophy type 2 (CGL2) [11]. Functional analysis revealed that the mutation results in a complete skipping of exon 6 leading to a predicted change in reading frame and an early termination of the protein. Clinical re-evaluation of the patients confirmed the diagnosis congenital generalized lipodystrophy.
Methods
Study subjects
We identified a consanguineous six generation Pakistani pedigree with three members, two females and one male, who presented with similar features. The pedigree structure suggested an autosomal recessive inheritance (Figure 1A). The family lived in an isolated rural area and the initial clinical data was scanty with information on muscular hypertrophy, mild mental retardation, and possible skeletal malformations. The diagnosis remained unclear and further clinical information and laboratory data were unavailable. Informed consent was obtained from all individuals who participated in this study or their legal guardian under a protocol approved by the local ethical committee at National Institute for Biotechnology and Genetic Engineering (NIBGE), Faisalabad, Pakistan. This includes consent from the patients to publish images.
Pedigree of family segregating Berardinelli-Seip congenital lipodystrophy and a BSCL2 splice site mutation. A) Family structure indicating consanguinity and autosomal recessive inheritance. Haplotypes spanning the BSCL2 gene mutation (c.574-2A > G) on chromosome 11 are shown below each symbol. The mutation segregates within a shared homozygous (autozygous) region in all affected individuals. B) The variant is predicted to lead to an exon skipping of exon 6, which would results in a frameshift and premature termination of the protein (p.Y256fsX48). C) Chromatogram showing cDNA sequences obtained from expressed mini-gene constructs containing BSCL1 gene spanning exons 5–7, with or without the variant. The variant is situated within the acceptor splice site of intron 5 of the BSCL2 gene. Wild-type (w.t.) BSCL1 construct (top), co-transfection of w.t. and mutant construct (middle) and mutated construct (bottom).
Exome sequencing and mutation detection
DNA from an affected individual (V3) was sonicated using a Covaris S2 instrument (Covaris, Inc., Woburn, MA, USA). Fragment libraries were created from the sheared samples using the AB Library Builder System (Life Technologies, Carlsbad, California, USA) and target enrichment was performed using the Agilent SureSelect Human All Exon v4 kit according to the manufacturer's protocols (Agilent, Santa Clara, CA, USA). Exome capture was conducted by hybridizing the DNA libraries with biotinylated RNA baits for 24 h followed by extraction using streptavidin coated magnetic beads. Captured DNA was then amplified followed by emulsion PCR using the EZ Bead System (Life Technologies) and sequenced on the SOLiD5500xl system, generating over 100 million reads of 75 bp length for each of the samples. Alignment of reads to the human reference sequence (hg19 assembly) and variant detection was performed using v2.1 of the LifeScope Software (Life Technologies). SNPs and indel data was stored in an in-house exome database together with variant annotation information obtained from ANNOVAR [12] and dbSNP137. Custom R scripts were used to identify potentially damaging variants not present in any of ~800 exomes from our in-house database. A minimal sequence depth of x5 was used as a quality cut-off. The BSCL2 variant was confirmed by direct sequencing with dideoxy chain-termination method (Applied Biosystems BigDye Terminator v3.1 Cycle Sequencing Kit) on a 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA). Sequence analysis was performed using the Sequencer software (Gene Codes Corporation, Ann Arbor, MI, USA).
Minigene expression construct
In total, 5 μg of genomic DNA from a heterozygous individual (VI:1) was used for amplification with primers BSCL2 e5F (GTG ATG CTG CAT TAC CGC TCA GAC) and BSCL2 e7R (CTG CAA AGA GAA GCG GTG TCG G). The PCR product was re-amplified using primers BSCL2 e5F Hind III (AGC TCA AGC TTC GTG ATG CTG CAT TAC CGC TCA GAC) and BSCL2 e7R Kpn I (CCC GCG GTA CCG CTG CAA AGA GAA GCG GTG TCG G) to introduce restriction sites. The resulting PCR product was cloned into a TA cloning vector (Invitrogen, Carlsbad, CA, USA) and individual clones were analyzed by Sanger sequencing. Clones representing the two alleles (wt allele (ag) and mutant allele (gg)) were identified and subsequently inserted into a pEGFP-C2 expression vector (Clontech, Mountain View, CA, USA) using Hind III, Kpn I and Rapid DNA ligation kit (Fermentas, Thermo Fisher Scientific, Waltham, MA, USA) and confirmed by sequencing. Thus, the minigene was fused to green fluorescent protein (Additional file 1: Figure S1A).
Splicing assay
The two minigene expression constructs were transfected into HEK293T cells using FuGene® Transfection reagent following manufacturer’s recommendations (Promega, Fitchburg, WI, USA). Cells were grown in DMEM supplemented with 10% fetal calf serum, 2 mM L-glutamine, 20 IU penicillin/streptomycin and non-essential amino acids (all Sigma-Aldrich, St. Louis, MI, USA) at 37°C with 5% CO2 in a humidified atmosphere. Expression of the GFP-minigene was confirmed by fluorescence microscopy after 48 hours. Subsequently, cells were harvested and total RNA was isolated using TRIZOLTM (Invitrogen), following the manufacturer’s protocols. Splicing of the minigene was detected by reverse transcription of 2 μg of total RNA using oligo-d(T) primers, utilizing the VILO cDNA synthesis kit (Invitrogen). Splicing products were amplified by standard PCR with primers Assay Forward (TAC AAG TCC GGC CGG ACT CAG ATC) and BSCL2 e7R (Additional file 1: Figure S1B). PCR products were cloned into a TA cloning vector (Invitrogen), and individual clones were analyzed by Sanger sequencing.
Results
Genetic analysis
Targeted enrichment of DNA from one affected family member, followed by WES and filtering, identified 34 homozygous variants, including one in the BSCL2 gene (NG_008461.1; NM_032667.6) (Additional file 2: Table S1). The sequencing resulted in 97% target base coverage (>1X) and 85% of the target bases were covered >20X. The variant is situated in the acceptor splice site of intron 5 (c.574-2A > G) of the gene and predicts skipping of exon 6 with a frameshift and premature termination codon (p.Y256fsX48) (Figure 1B). Sanger sequencing confirmed homozygosity for the c.574-2A > G variant in all three affected members (V:3, V:5 and VI:2) whereas three available parents were heterozygous (Figure 1A,C). Furthermore, the variant was excluded on 200 Swedish and 200 Pakistani control chromosomes and is not present in 6503 exomes from the Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), Seattle, WA (URL: http://evs.gs.washington.edu/EVS/). Segregation of a homozygous region surrounding the BSCL2 variant was confirmed using microsatellite markers (Figure 1A).
Study subjects
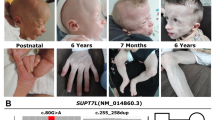
The family was revisited after WES analysis for a clinical re-evaluation of the three affected family members. The investigation revealed generalized lipodystrophy, axillary acanthosis nigricans relatively large hands and feet, muscular hypertrophy and acromegaloid appearance (Figure 2). The three affected individuals had mental retardation and individual V:5 had type 2 diabetes and a spastic gait suggesting upper motor neuron involvement. None of the three affected members had signs of muscle weakness. Hearing was normal. The proband (V:3) was available for ultrasound investigation and biochemical analysis. Ultrasound of abdomen showed enlarged liver (16.5 cm) and spleen (13.5 cm). Biochemical analysis of serum revealed increased levels of alanine aminotransferase levels (ALT; 64 U/L) and increased alkaline phosphatase levels (328 U/L). Unexpectedly, S-triglyceride and cholesterol levels were within the upper normal range as well as fasting glucose levels. The combined findings from our re-investigation of the family confirmed congenital generalized lipodystrophy.
Functional analysis
To evaluate the predicted effects of the acceptor splice site mutation in intron 5 of the BSCL2 gene, we created a mini-gene consisting of exon 5 through exon 7 including the two introns, fused to green fluorescent protein (Additional file 1: Figure S1A). Mini-genes containing the wild-type (wt; ag) and mutated (gg) splice site variants, respectively, were expressed in HEK293T cells and RNA was isolated 48 h after transfection. The amplified spliced products were analyzed by agarose gel electrophoresis and we observed a band of expected size from the wt construct (420 bp) whereas a shorter product (322 bp) was generated when amplifying from the mutated construct (Additional file 1: Figure S1B). The PCR products were cloned and analyzed by sequencing that confirmed the predicted skipping of exon 6 (Additional file 1: Figure S1C).
Discussion
We identified a consanguineous family segregating autosomal recessive muscular hypertrophy mild mental retardation and acromegaloid appearance in three individuals. Detailed clinical and biochemical data were unavailable and we decided to perform WES on a single affected family members in search for homozygous and potentially damaging gene variants. Analysis of WES data showed a homozygous acceptor splice site mutation, c.574-2A > G, in intron 5 of the BSCL2 gene. The BSCL2 gene is associated with congenital generalized lipodystrophy type 2 (CGL2) and, indeed, clinical re-evaluation of our patients confirmed characteristic clinical features of the disease in all three affected members [11]. Taken together the deleteriousness of the variant in BSCL2 and the clinical overlap with the preliminary information we had of the family, this was considered the most likely candidate variant of the ones indentified. Furthermore, we could by using a mini-gene construct spanning exons 5–7 of the mutated BSCL2 allele show that the mutation results in skipping of exon 6, an altered reading frame and a predicted early termination of the protein. Individuals with CGL2 can usually be recognized at birth, or soon thereafter, due to a near-total lack of body fat and prominent muscularity. Later characteristics include insulin resistance, hypertriglyceridemia, hepatic steatosis and early onset of diabetes. A high prevalence of mild to moderate intellectual impairment has been observed and patients have a higher risk for premature mortality, mainly due to cardiac failure. Additional features include acanthosis nigricans, hepatomegaly, hyperandrogenism and skeletal muscular hypertrophy [13, 14]. The disorder may, however, be mistaken for other types of lipodystrophies, especially in adult stage, making a clear diagnosis problematic [11, 15–18]. Considerable efforts have been made in the characterization and identification of the molecular basis of inherited lipodystrophies, and to date at least 11 gene-loci are known [13].
In our family, the initial information suggested a disabling condition segregating as an autosomal recessive trait. The clinical data were insufficient for a diagnosis and further investigations of the family were impossible within a reasonable time-frame. Thus, we decided to apply WES and, from our findings, we were able to suggest that the patients were affected by CGL2 within a few weeks from the sampling of DNA. Much later, the diagnosis was confirmed through directed clinical and biochemical investigations of the family. Moreover, this exemplifies a time-saving aspect of exome sequencing in situations of diagnostic challenges, inaccessible clinical data as well as acute illnesses [13]. Furthermore, rare diseases such as CGL2 may remain undiagnosed due to socio-economic constraints and inaccessible health care. Under such circumstances exome sequencing may become an attractive diagnostic complement with a continuous reduction in price. In a clinical setting, an alternative to WES could be the use of targeted sequencing of a specific subset of genes responsible for a certain condition. Although we are limited to clinically available disease-targeted enrichment tests, the resulting data generally have a much higher or often complete coverage of the targets [19].
Taken together, our results show the usefulness of WES for the identification of a single homozygous BSCL2 mutation in a family segregating an autosomal recessive trait. The mutation was analyzed in a cell-based assay that confirmed exon skipping. Using the findings from WES we could suggest the diagnosis of CGL2 in advance of complete clinical information from the patients. The use of exome sequencing accelerated and facilitated the subsequent and confirming investigations, with implications for care and follow-up of affected individuals.
Conclusions
This study illustrates how whole exome sequencing (WES) can be used in a clinical setting to identify gene mutations in search for a specific diagnosis. We show that WES can be used to accelerate clinical investigations of heterogeneous Mendelian traits for yet inconclusive cases.
Abbreviations
- BSCL:
-
Berardinelli-Seip congenital lipodystrophy
- CGL:
-
congenital generalized lipodystrophy.
References
Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, Nayir A, Bakkaloglu A, Ozen S, Sanjad S, Nelson-Williams C, Farhi A, Mane S, Lifton RP: Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A. 2009, 106 (45): 19096-19101.
Bonnefond A, Durand E, Sand O, De Graeve F, Gallina S, Busiah K, Lobbens S, Simon A, Bellanne-Chantelot C, Letourneau L, Scharfmann R, Delplanque J, Sladek R, Polak M, Vaxillaire M, Froguel P: Molecular diagnosis of neonatal diabetes mellitus using next-generation sequencing of the whole exome. PLoS One. 2010, 5 (10): e13630-
Montenegro G, Powell E, Huang J, Speziani F, Edwards YJ, Beecham G, Hulme W, Siskind C, Vance J, Shy M, Zuchner S: Exome sequencing allows for rapid gene identification in a Charcot-Marie-Tooth family. Ann Neurol. 2011, 69 (3): 464-470.
Pierson TM, Adams DA, Markello T, Golas G, Yang S, Sincan M, Simeonov DR, Fuentes Fajardo K, Hansen NF, Cherukuri PF, Cruz P, Teer JK, Mullikin JC, Boerkoel CF, Gahl WA, Tifft CJ: Exome sequencing as a diagnostic tool in a case of undiagnosed juvenile-onset GM1-gangliosidosis. Neurology. 2012, 79 (2): 123-126.
Dias C, Sincan M, Cherukuri PF, Rupps R, Huang Y, Briemberg H, Selby K, Mullikin JC, Markello TC, Adams DR, Gahl WA, Boerkoel CF: An analysis of exome sequencing for diagnostic testing of the genes associated with muscle disease and spastic paraplegia. Hum Mutat. 2012, 33 (4): 614-626.
Futema M, Plagnol V, Whittall RA, Neil HA, Humphries SE: Use of targeted exome sequencing as a diagnostic tool for Familial Hypercholesterolaemia. J Med Genet. 2012, 49 (10): 644-649.
de Ligt J, Willemsen MH, van Bon BW, Kleefstra T, Yntema HG, Kroes T, Vulto-van Silfhout AT, Koolen DA, de Vries P, Gilissen C, del Rosario M, Hoischen A, Scheffer H, de Vries BB, Brunner HG, Veltman JA, Vissers LE: Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012, 367 (20): 1921-1929.
Hammer MB, Eleuch-Fayache G, Gibbs JR, Arepalli SK, Chong SB, Sassi C, Bouhlal Y, Hentati F, Amouri R, Singleton AB: Exome sequencing: an efficient diagnostic tool for complex neurodegenerative disorders. Eur J Neurol. 2013, 20 (3): 486-492.
Tsurusaki Y, Kobayashi Y, Hisano M, Ito S, Doi H, Nakashima M, Saitsu H, Matsumoto N, Miyake N: The diagnostic utility of exome sequencing in Joubert syndrome and related disorders. J Hum Genet. 2013, 58 (2): 113-115.
Hanchard NA, Murdock DR, Magoulas PL, Bainbridge M, Muzny D, Wu YQ, Wang M, McGuire AL, Lupski JR, Gibbs RA, Brown CW: Exploring the utility of whole-exome sequencing as a diagnostic tool in a child with atypical episodic muscle weakness. Clin Genet. 2013, 83 (5): 457-461.
Magre J, Delepine M, Khallouf E, Gedde-Dahl T, Van Maldergem L, Sobel E, Papp J, Meier M, Megarbane A, Bachy A, Verloes A, d'Abronzo FH, Seemanova E, Assan R, Baudic N, Bourut C, Czernichow P, Huet F, Grigorescu F, de Kerdanet M, Lacombe D, Labrune P, Lanza M, Loret H, Matsuda F, Navarro J, Nivelon-Chevalier A, Polak M, Robert JJ, Tric P, et al: Identification of the gene altered in Berardinelli-Seip congenital lipodystrophy on chromosome 11q13. Nat Genet. 2001, 28 (4): 365-370.
Wang K, Li M, Hakonarson H: ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38 (16): e164-
Garg A: Clinical review#: Lipodystrophies: genetic and acquired body fat disorders. J Clin Endocrinol Metab. 2011, 96 (11): 3313-3325.
Van Maldergem L, Magre J, Khallouf TE, Gedde-Dahl T, Delepine M, Trygstad O, Seemanova E, Stephenson T, Albott CS, Bonnici F, Panz VR, Medina JL, Bogalho P, Huet F, Savasta S, Verloes A, Robert JJ, Loret H, De Kerdanet M, Tubiana-Rufi N, Megarbane A, Maassen J, Polak M, Lacombe D, Kahn CR, Silveira EL, D'Abronzo FH, Grigorescu F, Lathrop M, Capeau J, et al: Genotype-phenotype relationships in Berardinelli-Seip congenital lipodystrophy. J Med Genet. 2002, 39 (10): 722-733.
Agarwal AK, Arioglu E, De Almeida S, Akkoc N, Taylor SI, Bowcock AM, Barnes RI, Garg A: AGPAT2 is mutated in congenital generalized lipodystrophy linked to chromosome 9q34. Nat Genet. 2002, 31 (1): 21-23.
Hayashi YK, Matsuda C, Ogawa M, Goto K, Tominaga K, Mitsuhashi S, Park YE, Nonaka I, Hino-Fukuyo N, Haginoya K, Sugano H, Nishino I: Human PTRF mutations cause secondary deficiency of caveolins resulting in muscular dystrophy with generalized lipodystrophy. J Clin Invest. 2009, 119 (9): 2623-2633.
Kim CA, Delepine M, Boutet E, El Mourabit H, Le Lay S, Meier M, Nemani M, Bridel E, Leite CC, Bertola DR, Semple RK, O'Rahilly S, Dugail I, Capeau J, Lathrop M, Magre J: Association of a homozygous nonsense caveolin-1 mutation with Berardinelli-Seip congenital lipodystrophy. J Clin Endocrinol Metab. 2008, 93 (4): 1129-1134.
Windpassinger C, Auer-Grumbach M, Irobi J, Patel H, Petek E, Horl G, Malli R, Reed JA, Dierick I, Verpoorten N, Warner TT, Proukakis C, Van den Bergh P, Verellen C, Van Maldergem L, Merlini L, De Jonghe P, Timmerman V, Crosby AH, Wagner K: Heterozygous missense mutations in BSCL2 are associated with distal hereditary motor neuropathy and Silver syndrome. Nat Genet. 2004, 36 (3): 271-276.
Rehm HL: Disease-targeted sequencing: a cornerstone in the clinic. Nat Rev Genet. 2013, 14 (4): 295-300.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2350/15/71/prepub
Acknowledgements
Supported in part by the Swedish Research Council (K2010-66X-10829-17-3 and 621-2009-4629) and Swedish Links, Asia (348-2008-6069), Science for Life laboratory, Uppsala University Hospital and Uppsala University. J.K is supported by the Swedish Society for Medical Research. We thank all family members who participated in this study. We also thank the Uppsala Genome Center for technical support.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JS helped in the design of the study, carried out the functional analyses and helped to draft the manuscript. TNK, MT, PA and KM carried out the molecular genetic studies. SMB and TNK carried out the clinical evaluation of the patients. JK designed the study and drafted the manuscript. All authors read and approved the final manuscript.
Electronic supplementary material
12881_2014_1215_MOESM1_ESM.png
Additional file 1: Figure S1: A) Minigene construct consisting of exon 5 through to exon 7 of the BSCL2 gene, including the introns, fused to green fluorescent protein (GFP). Two minigenes representing wt (ag) and mutated (gg) splice site were used. B) PCR based assay of the minigenes. An observed band of expected size in the wt construct (420 bp), but a smaller product (322 bp) corresponding to the expected exon skipping could be identified from the mutated construct. C) Sanger sequencing of cDNA from mutated (gg) construct confirms skipping of exon 6. (PNG 625 KB)
12881_2014_1215_MOESM2_ESM.docx
Additional file 2: Table S1: Homozygous variants identified by WES. *snp137 indicates if the variant is present in the 137 version of dbSNP. Note that presented variants, besides the identified variant in the BSCL2 gene, are not confirmed by Sanger sequencing, and could thereby possibly be sequencing errors. (DOCX 16 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Schuster, J., Khan, T.N., Tariq, M. et al. Exome sequencing circumvents missing clinical data and identifies a BSCL2 mutation in congenital lipodystrophy. BMC Med Genet 15, 71 (2014). https://doi.org/10.1186/1471-2350-15-71
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2350-15-71