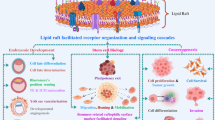
Abstract
Lipid rafts are the lateral assemblies of cholesterol, sphingomyelin, glycosphingolipids, and specific proteins within the cell plasma membrane. These microdomains are involved in a number of important cellular processes including membrane rearrangement, protein internalization, signal transduction, and the entry of viruses into a cell. Some of the lipid rafts are stabilized by special microdomain-forming proteins such as caveolins, SPFH domain-containing superfamily, tetraspanins, galectins, which maintain the integrity of rafts and regulate many resident proteins, thereby participating in nearly all life processes of cells. However, such classes of microdomain-forming proteins are still considered separately from each other. In this review we have tried to perform a complex analysis of microdomain-forming proteins in cell regulation by the example of EGFR receptors, integrins, and matrix metalloproteinases.
Similar content being viewed by others
References
Nicolson G.L., Singer S.J. 1974. The distribution and asymmetry of mammalian cell surface saccharides utilizing ferritin-conjugated plant agglutinins as specific saccharide stains. J. Cell Biol. 60, 236–248.
Simons K., Sampaio J.L. 2011. Membrane organization and lipid rafts. Cold Spring Harb. Perspect. Biol. 3, a004697.
Lindner R., Naim H.Y. 2009. Domains in biological membranes. Exp. Cell Res. 315, 2871–2878.
Kirkham M., Nixon S.J., Howes M.T., Abi-Rached L., Wakeham D.E., Hanzal-Bayer M., Ferguson C., Hill M.M., Fernandez-Rojo M., Brown D.A., Hancock J.F., Brodsky F.M., Parton R.G. 2008. Evolutionary analysis and molecular dissection of caveola biogenesis. J. Cell Sci. 121, 2075–2086.
Razani B., Woodman S.E., Lisanti M.P. 2002. Caveolae: From cell biology to animal physiology. Pharmacol. Rev. 54, 431–467.
Williams T.M., Lisanti M.P. 2004. The caveolin proteins. Genome Biol. 5, 214.
Lisanti M.P., Scherer P.E., Tang Z., Sargiacomo M. 1994. Caveolae, caveolin and caveolin-rich membrane domains: A signalling hypothesis. Trends Cell Biol. 4, 231–235.
Bastiani M., Parton R.G. 2010. Caveolae at a glance. J. Cell Sci. 123, 3831–3836.
Ishikawa Y., Otsu K., Oshikawa J. 2005. Caveolin; different roles for insulin signal? Cell Signal. 17, 1175–1182.
Le Lay S., Blouin C.M., Hajduch E., Dugail I. 2009. Filling up adipocytes with lipids. Lessons from caveolin-1 deficiency. Biochim. Biophys. Acta. 1791, 514–518.
Huang S., Tian H., Chen Z., Yu T., Xu A. 2010. The evolution of vertebrate tetraspanins: Gene loss, retention, and massive positive selection after whole genome duplications. BMC Evol. Biol. 10, 306.
Levy S., Shoham T. 2005. Protein-protein interactions in the tetraspanin web. Physiology. 20, 218–224.
Yanez-Mo M., Barreiro O., Gordon-Alonso M., Sala-Valdes M., Sanchez-Madrid F. 2009. Tetraspaninenriched microdomains: A functional unit in cell plasma membranes. Trends Cell Biol. 19, 434–446.
Rivera-Milla E., Stuermer C.A., Malaga-Trillo E. 2006. Ancient origin of reggie (flotillin), reggie-like, and other lipid-raft proteins: Convergent evolution of the SPFH domain. Cell Mol. Life Sci. 63, 343–357.
Browman D.T., Hoegg M.B., Robbins S.M. 2007. The SPFH domain-containing proteins: More than lipid raft markers. Trends Cell Biol. 17, 394–402.
Babuke T., Tikkanen R. 2007. Dissecting the molecular function of reggie/flotillin proteins. Eur. J. Cell Biol. 86, 525–532.
Stuermer C.A. 2010. The reggie/flotillin connection to growth. Trends Cell Biol. 20, 6–13.
Frick M., Bright N.A., Riento K., Bray A., Merrified C., Nichols B.J. 2007. Coassembly of flotillins induces formation of membrane microdomains, membrane curvature, and vesicle budding. Curr. Biol. 17, 1151–1156.
Babuke T., Ruonala M., Meister M., Amaddii M., Genzler C., Esposito A., Tikkanen R. 2009. Heterooligomerization of reggie-1/flotillin-2 and reggie-2/flotillin-1 is required for their endocytosis. Cell Signal. 21, 1287–1297.
Aït-Slimane T., Galmes R., Trugnan G., Maurice M. 2009. Basolateral internalization of GPI-anchored proteins occurs via a clathrin-independent flotillindependent pathway in polarized hepatic cells. Mol. Biol. Cell. 20, 3792–3800.
Lajoie P., Goetz J.G., Dennis J.W., Nabi I.R. 2009. Lattices, rafts, and scaffolds: Domain regulation of receptor signaling at the plasma membrane. J. Cell Biol. 185, 381–385.
Boscher C., Dennis J.W., Nabi I.R. 2011. Glycosylation, galectins and cellular signaling. Curr. Opin. Cell Biol. 23, 383–392.
Williams T.M., Lisanti M.P. 2004. The Caveolin genes: From cell biology to medicine. Ann. Med. 36, 584–595.
Mercier I., Jasmin J.F., Pavlides S., Minetti C., Flomenberg N., Pestell R.G., Frank P.G., Sotgia F., Lisanti M.P. 2009. Clinical and translational implications of the caveolin gene family: Lessons from mouse models and human genetic disorders. Lab. Invest. 89, 614–623.
Cassoni P., Daniele L., Maldi E., Righi L., Tavaglione V., Novello S., Volante M., Scagliotti G.V., Papotti M. 2009. Caveolin-1 expression in lung carcinoma varies according to tumour histotype and is acquired de novo in brain metastases. Histopathology. 55, 20–27.
Wiechen K., Diatchenko L., Agoulnik A., Scharff K.M., Schober H., Arlt K., Zhumabayeva B., Siebert P.D., Dietel M., Schäfer R., Sers C. 2001. Caveolin-1 is down-regulated in human ovarian carcinoma and acts as a candidate tumor suppressor gene. Am. J. Pathol. 159, 1635–1643.
Bender F.C., Reymond M.A., Bron C., Quest A.F.G. 2000. Caveolin-1 levels are down-regulated in human colon tumors, and ectopic expression of caveolin-1 in colon carcinoma cell lines reduces cell tumorigenicity. Cancer Res. 60, 5870–5878.
Hino M., Doihara H., Kobayashi K., Aoe M., Shimizu N. 2003. Caveolin-1 as tumor suppressor gene in breast cancer. Surg. Today. 33, 486–490.
Wiechen K., Sers C., Agoulnik A., Arlt K., Dietel M., Schlag P.M., Schneider U. 2001. Down-regulation of caveolin-1, a candidate tumor suppressor gene, in sarcomas. Am. J. Pathol. 158, 833–839.
Bayer-Garner I., Morgan M., Smoller B.R. 2002. Caveolin expression is common among benign and malignant smooth muscle and adipocyte neoplasms. Mod. Pathol. 1, 1–5.
Arkhipova K.A., Rybko V.A., Zemlyakova V.V., Bliznyukov O.P., Martynkov D.V., Gubina G.I., Zborovskaya I.B. 2009. Caveolin-1 expression in soft tissue tumors. J. N.N. Blokhin RCRS RAMS (Rus.). 1, 4–9.
Colnot C., Fowlis D., Ripoche M.A., Bouchaert I., Poirier F. 1998. Embryonic implantation in galectin 1/ galectin 3 double mutant mice. Dev. Dyn. 211, 306–313.
Newlaczyl A.U., Yu L.G. 2011. Galectin-3 — a jack-of-all-trades in cancer. Cancer Lett. 313, 123–128.
Nangia-Makker P., Balan V., Raz A. 2008. Regulation of tumor progression by extracellular galectin-3. Cancer Microenviron. 1, 43–51.
Dumic J., Dabelic S., Flögel M. Galectin-3: An openended story. Biochim. Biophys. Acta. 1760, 616–635.
Knobeloch K.P., Wright M.D., Ochsenbein A.F., Liesenfeld O., Löhler J., Zinkernagel R.M., Horak I., Orinska Z. 2000. Targeted inactivation of the tetraspanin CD37 impairs T-cell-dependent B-cell response under suboptimal costimulatory conditions. Mol. Cell Biol. 220, 5363–5369.
Miyazaki T., Müller U., Campbell K.S. 1997. Normal development but differentially altered proliferative responses of lymphocytes in mice lacking CD81. EMBO J. 16, 4217–4225.
van Soest S., Westerveld A., de Jong P.T., Bleeker-Wagemakers E.M., Bergen A.A. 1999. Retinitis pigmentosa: Defined from a molecular point of view. Surv. Ophthalmol. 43, 321–334.
Le Naour F., Rubinstein E., Jasmin C., Prenant M., Boucheix C. 2000. Severely reduced female fertility in CD9-deficient mice. Science. 287, 319–321.
Miyado K., Yamada G., Yamada S., Hasuwa H., Nakamura Y., Ryu F., Suzuki K., Kosai K., Inoue K., Ogura A., Okabe M., Mekada E. 2000. Requirement of CD9 on the egg plasma membrane for fertilization. Science. 287, 321–324.
Zemni R., Bienvenu T., Vinet M.C., Sefiani A., Carrié A., Billuart P., McDonell N., Couvert P., Francis F., Chafey P., Fauchereau F., Friocourt G., des Portes V., Cardona A., Frints S., Meindl A., Brandau O., Ronce N., Moraine C., van Bokhoven H., Ropers H.H., Sudbrak R., Kahn A., Fryns J.P., Beldjord C., Chelly J. 2000. A new gene involved in X-linked mental retardation identified by analysis of an X;2 balanced translocation. Nat. Genet. 24, 167–170.
Hazarika P., McCarty M.F., Prieto V.G., George S., Babu D., Koul D., Bar-Eli M., Duvic M. 2004. Upregulation of Flotillin-2 is associated with melanoma progression and modulates expression of the thrombin receptor protease activated receptor 1. Cancer Res. 64, 7361–7369.
Lin C., Wu Z., Lin X., Yu C., Shi T., Zeng Y., Wang X., Li J., Song L. 2011. Knockdown of FLOT1 impairs cell proliferation and tumorigenicity in breast cancer through upregulation of FOXO3a. Clin. Cancer Res. 17, 3089–3099.
Morrow I.C., Parton R.G. 2005. Flotillins and the PHB domain protein family: Rafts, worms and anaesthetics. Traffic. 6, 725–740.
Langhorst M.F., Reuter A., Stuermer C.A. 2005. Scaffolding microdomains and beyond: The function of reggie/flotillin proteins. Cell Mol. Life Sci. 62, 2228–2240.
Jang I.H., Kim J.H., Lee B.D., Bae S.S., Park M.H., Suh P.G., Ryu S.H. 2001. Localization of phospholipase C-gamma1 signaling in caveolae: Importance in EGF-induced phosphoinositide hydrolysis but not in tyrosine phosphorylation. FEBS Lett. 491, 4–8.
Couet J., Sargiacomo M., Lisanti M.P. 1997. Interaction of a receptor tyrosine kinase, EGF-R, with caveolins. Caveolin binding negatively regulates tyrosine and serine/threonine kinase activities. J. Biol. Chem. 272, 30429–30438.
Mineo C., James G.L., Smart E.J., Anderson R.G. 1996. Localization of epidermal growth factor-stimulated Ras/Raf-1 interaction to caveolae membrane. J. Biol. Chem. 271, 11930–11935.
Mineo C., Gill G.N., Anderson R.G. 1999. Regulated migration of epidermal growth factor receptor from caveolae. J. Biol. Chem. 274, 30636–30643.
Glenney J.R. Jr., Zokas L. 1989. Novel tyrosine kinase substrates from Rous sarcoma virus-transformed cells are present in the membrane skeleton. J. Cell Biol. 108, 2401–2408.
Lee H., Volonte D., Galbiati F., Iyengar P., Lublin D.M., Bregman D.B., Wilson M.T., Campos-Gonzalez R., Bouzahzah B., Pestell R.G., Scherer P.E., Lisanti M.P. 2000. Constitutive and growth factor-regulated phosphorylation of caveolin-1 occurs at the same site (Tyr-14) in vivo: Identification of a c-Src/Cav-1/Grb7 signaling cassette. Mol. Endocrinol. 14, 1750–1775.
Cao H., Courchesne W.E., Mastick C.C. 2002. A phosphotyrosine-dependent protein interaction screen reveals a role for phosphorylation of caveolin-1 on tyrosine 14: Recruitment of C-terminal Src kinase. J. Biol. Chem. 277, 8771–8774.
Galbiati F., Volonte D., Engelman J.A., Watanabe G., Burk R., Pestell R.G., Lisanti M.P. 1998. Targeted downregulation of caveolin-1 is sufficient to drive cell transformation and hyperactivate the p42/44 MAP kinase cascade. EMBO J. 17, 6633–6648.
Park W.Y., Park J.S., Cho K.A., Kim D.I., Ko Y.G., Seo J.S., Park S.C. 2000. Up-regulation of caveolin attenuates epidermal growth factor signaling in senescent cells. J. Biol. Chem. 275, 20847–20852.
Engelman J.A., Zhang X.L., Razani B., Pestell R.G., Lisanti M.P. 1999. p42/44 MAP kinase-dependent and -independent signaling pathways regulate caveolin-1 gene expression. Activation of Ras-MAP kinase and protein kinase a signaling cascades transcriptionally down-regulates caveolin-1 promoter activity. J. Biol. Chem. 274, 32333–32341.
Khan E.M., Heidinger J.M., Levy M., Lisanti M.P., Ravid T., Goldkorn T. 2006. Epidermal growth factor receptor exposed to oxidative stress undergoes Src- and caveolin-1-dependent perinuclear trafficking. J. Biol. Chem. 281, 14486–14493.
Lajoie P., Partridge E.A., Guay G., Goetz J.G., Pawling J., Lagana A., Joshi B., Dennis J.W., Nabi I.R. 2007. Plasma membrane domain organization regulates EGFR signaling in tumor cells. J. Cell. Biol. 179, 341–356.
Roepstorff K., Thomsen P., Sandvig K., van Deurs B. 2002. Sequestration of epidermal growth factor receptors in non-caveolar lipid rafts inhibits ligand binding. J. Biol. Chem. 277, 18954–18960.
Matveev S.V., Smart E.J. 2002. Heterologous desensitization of EGF receptors and PDGF receptors by sequestration in caveolae. Am. J. Physiol. Cell Physiol. 282, C935–946.
Pike L.J. 2005. Growth factor receptors, lipid rafts and caveolae: An evolving story. Biochim. Biophys. Acta. 1746, 260–273.
de Laurentiis A., Donovan L., Arcaro A. 2007. Lipid rafts and caveolae in signaling by growth factor receptors. Open Biochem. J. 1, 12–32.
Murayama Y., Shinomura Y., Oritani K., Miyagawa J., Yoshida H., Nishida M., Katsube F., Shiraga M., Miyazaki T., Nakamoto T., Tsutsui S., Tamura S., Higashiyama S., Shimomura I., Hayashi N. 2008. The tetraspanin CD9 modulates epidermal growth factor receptor signaling in cancer cells. J. Cell Physiol. 216, 135–143.
Chen S., Sun Y., Jin Z., Jing X. 2011. Functional and biochemical studies of CD9 in fibrosarcoma cell line. Mol. Cell Biochem. 350, 89–99.
Odintsova E., Sugiura T., Berditchevski F. 2000. Attenuation of EGF receptor signaling by a metastasis suppressor, the tetraspanin CD82/KAI-1. Curr. Biol. 10, 1009–1012.
Odintsova E., Voortman J., Gilbert E., Berditchevski F. 2003. Tetraspanin CD82 regulates compartmentalisation and ligand-induced dimerization of EGFR. J. Cell Sci. 116, 4557–4566.
Wang X.Q., Yan Q., Sun P., Liu J.W., Go L., McDaniel S.M., Paller A.S. 2007. Suppression of epidermal growth factor receptor signaling by protein kinase C-alpha activation requires CD82, caveolin-1, and ganglioside. Cancer Res. 67, 9986–9995.
Zhang X.A., Bontrager A.L., Hemler M.E. 2001. Transmembrane-4 superfamily proteins associate with activated protein kinase C (PKC) and link PKC to specific beta(1) integrins. J. Biol. Chem. 276, 25005–25013.
Kazarov A.R., Yang X., Stipp C.S., Sehgal B., Hemler M.E. 2002. An extracellular site on tetraspanin CD151 determines alpha 3 and alpha 6 integrindependent cellular morphology. J. Cell Biol. 158, 1299–1309.
Mannion B.A., Berditchevski F., Kraeft S.K., Chen L.B., Hemler M.E. 1996. Transmembrane-4 superfamily proteins CD81 (TAPA-1), CD82, CD63, and CD53 specifically associated with integrin alpha 4 beta 1 (CD49d/CD29). J. Immunol. 157, 2039–2047.
Berditchevski F. 2001. Complexes of tetraspanins with integrins: More than meets the eye. J. Cell Sci. 114, 4143–4151.
Yang X., Wei L.L., Tang C., Slack R., Mueller S., Lippman M.E. 2001. Overexpression of KAI1 suppresses in vitro invasiveness and in vivo metastasis in breast cancer cells. Cancer Res. 61, 5284–5288.
Hong I.K., Jin Y.J., Byun H.J., Jeoung D.I., Kim Y.M., Lee H. 2006. Homophilic interactions of Tetraspanin CD151 up-regulate motility and matrix metalloproteinase-9 expression of human melanoma cells through adhesion-dependent c-Jun activation signaling pathways. J. Biol. Chem. 281, 24279–24292.
Larsson C. 2006. Protein kinase C and the regulation of the actin cytoskeleton. Cell Signal. 18, 276–284.
Miranti C.K. 2009. Controlling cell surface dynamics and signaling: How CD82/KAI1 suppresses metastasis. Cell Signal. 21, 196–211.
Zhang X.A., He B., Zhou B., Liu L. 2003. Requirement of the p130CAS-Crk coupling for metastasis suppressor KAI1/CD82-mediated inhibition of cell migration. J. Biol. Chem. 278, 27319–27328.
Delaguillaumie A., Lagaudriere-Gesbert C., Popoff M.R., Conjeaud H. 2002. Rho GTPases link cytoskeletal rearrangements and activation processes induced via the tetraspanin CD82 in T lymphocytes. J. Cell Sci. 115, 433–443.
Johnson J.L., Winterwood N., DeMali K.A., Stipp C.S. 2009. Tetraspanin CD151 regulates RhoA activation and the dynamic stability of carcinoma cell-cell contacts. J. Cell Sci. 122, 2263–2273.
Baldwin G., Novitskaya V., Sadej R., Pochec E., Litynska A., Hartmann C., Williams J., Ashman L., Eble J.A., Berditchevski F. 2008. Tetraspanin CD151 regulates glycosylation of (alpha)3(beta)1 integrin. J. Biol. Chem. 283, 35445–35454.
Nishiuchi R., Sanzen N., Nada S., Sumida Y., Wada Y., Okada M., Takagi J., Hasegawa H., Sekiguchi K. 2005. Potentiation of the ligand-binding activity of integrin alpha3beta1 via association with tetraspanin CD151. Proc. Natl. Acad. Sci. USA. 102, 1939–1944.
Hasegawa M., Furuya M., Kasuya Y., Nishiyama M., Sugiura T., Nikaido T., Momota Y., Ichinose M., Kimura S. 2007. CD151 dynamics in carcinomastroma interaction: Integrin expression, adhesion strength and proteolytic activity. Lab. Invest. 87, 882–892.
Liu L., He B., Liu W.M., Zhou D., Cox J.V., Zhang X.A. 2007. Tetraspanin CD151 promotes cell migration by regulating integrin trafficking. J. Biol. Chem. 282, 31631–31642.
Parat M.-O., Anand-Apte B., Fox P.L. 2003. Differential Caveolin-1 polarization in endothelial cells during migration in two and three dimensions. Mol. Biol. Cell. 14, 3156–3168.
Beardsley A., Fang K., Mertz H., Castranova V., Friend S., Liu J. 2005. Loss of Caveolin-1 polarity impedes endothelial cell polarization and directional movement. J. Biol. Chem. 280, 3541–3547.
Grande-Garcia A., Echarri A., de Rooij J., Alderson N.B., Waterman-Storer C.M., Valdivielso J.M., del Pozo M.A. 2007. Caveolin-1 regulates cell polarization and directional migration through Src kinase and Rho GTPases. J. Cell Biol. 177, 683–694.
Wei Y., Yang X., Liu Q., Wilkins J.A., Chapman H.A. 1999. A role for caveolin and the urokinase receptor in integrin-mediated adhesion and signaling. J. Cell Biol. 144, 1285–1294.
Goetz J.G., Joshi B., Lajoie P., Strugnell S.S., Scudamore T., Kojic L.D., Nabi I.R. 2008. Concerted regulation of focal adhesion dynamics by galectin-3 and tyrosine-phosphorylated caveolin-1. J. Cell Biol. 180, 1261–1275.
Bass R., Werner F., Odintsova E., Sugiura T., Berditchevski F., Ellis V. 2005. Regulation of urokinase receptor proteolytic function by the tetraspanin CD82. J. Biol. Chem. 280, 14811–14818.
Langhorst M.F., Jaegera F.A., Mueller S., Sven Hartmann L., Luxenhofer G., Stuermer C.A. 2008. Reggies/flotillins regulate cytoskeletal remodeling during neuronal differentiation via CAP/ponsin and Rho GTPases. Eur. J. Cell Biol. 87, 921–931.
Lypez-Casas P.P., del Mazo J. 2003. Regulation of flotillin-1 in the establishment of NIH-3T3 cell-cell interactions. FEBS Lett. 555, 223–228.
Gialeli C., Theocharis A.D., Karamanos N.K. 2011. Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. FEBS J. 278, 16–27.
Yamaguchi H., Takeo Y., Yoshida S., Kouchi Z., Nakamura Y., Fukami K. 2009. Lipid rafts and caveolin-1 are required for invadopodia formation and extracellular matrix degradation by human breast cancer cells. Cancer Res. 69, 8594–8602.
Kim H.N., Chung H.S. 2008. Caveolin-1 inhibits membrane-type 1 matrix metalloproteinase activity. BMB Rep. 41, 858–862.
Williams T.M., Medina F., Badano I., Hazan R.B., Hutchinson J., Muller W.J., Chopra N.G., Scherer P.E., Pestell R.G., Lisanti M.P. 2004. Caveolin-1 gene disruption promotes mammary tumorigenesis and dramatically enhances lung metastasis in vivo. Role of Cav-1 in cell invasiveness and matrix metalloproteinase (MMP-2/9) secretion. J. Biol. Chem. 279, 51630–51646.
Han F., Zhu H.G. 2010. Caveolin-1 regulating the invasion and expression of matrix metalloproteinase (MMPs) in pancreatic carcinoma cells. J. Surg. Res. 159, 443–450.
Jia L., Wang S., Zhou H., Cao J., Hu Y., Zhang J. 2006. Caveolin-1 up-regulates CD147 glycosylation and the invasive capability of murine hepatocarcinoma cell lines. Int. J. Biochem. Cell Biol. 38, 1584–1593.
Tang W., Hemler M.E. 2004. Caveolin-1 regulates matrix metalloproteinases-1 induction and CD147/ EMMPRIN cell surface clustering. J. Biol. Chem. 279, 11112–11118.
Yanez-Mo M., Barreiro O., Gonzalo P., Batista A., Megías D., Genís L., Sachs N., Sala-Valdes M., Alonso M.A., Montoya M.C., Sonnenberg A., Arroyo A.G., Sanchez-Madrid F. 2008. MT1-MMP collagenolytic activity is regulated through association with tetraspanin CD151 in primary endothelial cells. Blood. 112, 3217–3226.
Lafleur M.A., Xu D., Hemler M.E. 2009. Tetraspanin proteins regulate membrane type-1 matrix metalloproteinase-dependent pericellular proteolysis. Mol. Biol. Cell. 20, 2030–2040.
Saito Y., Tachibana I., Takeda Y., Yamane H., He P., Suzuki M., Minami S., Kijima T., Yoshida M., Kumagai T., Osaki T., Kawase I. 2006. Absence of CD9 enhances adhesion-dependent morphologic differentiation, survival, and matrix metalloproteinase-2 production in small cell lung cancer cells. Cancer Res. 66, 9557–9565.
Hong I.K., Kim Y.M., Jeoung D.I., Kim K.C., Lee H. 2005. Tetraspanin CD9 induces MMP-2 expression by activating p38 MAPK, JNK and c-Jun pathways in human melanoma cells. Exp. Mol. Med. 37, 230–239.
Park J.E., Chang W.Y., Cho M. 2009. Induction of matrix metalloproteinase-9 by galectin-7 through p38 MAPK signaling in HeLa human cervical epithelial adenocarcinoma cells. Oncol. Rep. 22, 1373–1379.
Demers M., Magnaldo T., St-Pierre Y. 2005. A novel function for galectin-7: promoting tumorigenesis by up-regulating MMP-9 gene expression. Cancer Res. 65, 5205–5210.
Wu M.H., Hong T.M., Cheng H.W., Pan S.H., Liang Y.R., Hong H.C., Chiang W.F., Wong T.Y., Shieh D.B., Shiau A.L., Jin Y.T., Chen Y.L. 2009. Galectin-1-mediated tumor invasion and metastasis, up-regulated matrix metalloproteinase expression, and reorganized actin cytoskeletons. Mol. Cancer Res. 7, 311–318.
Prudova A., auf dem Keller U., Butler G.S., Overall C.M. 2010. Multiplex N-terminome analysis of MMP-2 and MMP-9 substrate degradomes by iTRAQ-TAILS quantitative proteomics. Mol. Cell Proteomics. 9, 894–911.
Dean R.A., Overall C.M. 2007. Proteomics discovery of metalloproteinase substrates in the cellular context by iTRAQ labeling reveals a diverse MMP-2 substrate degradome. Mol. Cell Proteomics. 6, 611–623.
Ochieng J., Fridman R., Nangia-Makker P., Kleiner D.E., Liotta L.A., Stetler-Stevenson W.G., Raz A. 1994. Galectin-3 is a novel substrate for human matrix metalloproteinases-2 and -9. Biochemistry. 33, 14109–14114.
Ochieng J., Green B., Evans S., James O., Warfield P. 1998. Modulation of the biological functions of galectin-3 by matrix metalloproteinases. Biochim. Biophys. Acta. 1379, 97–106.
Bist A., Fielding C.J., Fielding P.E. 2000. p53 regulates caveolin gene transcription, cell cholesterol, and growth by a novel mechanism. Biochemistry. 39, 1966–1972.
Gaudin J.C., Arar C., Monsigny M., Legrand A. 1997. Modulation of the expression of the rabbit galectin-3 gene by p53 and c-Ha-ras proteins and PMA. Glycobiology. 7, 1089–1098.
Dumic J., Lauc G., Flogel M. 2000. Expression of galectin-3 in cells exposed to stress-roles of jun and NF-kappaB. Cell Physiol. Biochem. 10, 149–158.
Deregowski V., Delhalle S., Benoit V., Bours V., Merville M.P. 2002. Identification of cytokine-induced nuclear factor-kappaB target genes in ovarian and breast cancer cells. Biochem. Pharmacol. 64, 873–881.
Li J., Peet G.W., Balzarano D., Li X., Massa P., Barton R.W., Marcu K.B. 2001. Novel NEMO/IkappaB kinase and NF-kappa B target genes at the pre-B to immature B cell transition. J. Biol. Chem. 276, 18579–18590.
Dong J.T., Isaacs W.B., Barrett J.C., Isaacs J.T. 1997. Genomic organization of the human KAI1 metastasissuppressor gene. Genomics. 41, 25–32.
Sasaki Y., Oshima Y., Koyama R., Maruyama R., Akashi H., Mita H., Toyota M., Shinomura Y., Imai K., Tokino T. 2008. Identification of flotillin-2, a major protein on lipid rafts, as a novel target of p53 family members. Mol. Cancer Res. 6, 395–406.
Wang J., Liu X., Ni P., Gu Z., Fan Q. 2010. SP1 is required for basal activation and chromatin accessibility of CD151 promoter in liver cancer cells. Biochem. Biophys. Res. Commun. 393, 291–296.
Cao S., Fernandez-Zapico M.E., Jin D., Puri V., Cook T.A., Lerman L.O., Zhu X.Y., Urrutia R., Shah V. 2005. KLF11-mediated repression antagonizes Sp1/sterol-responsive element-binding proteininduced transcriptional activation of caveolin-1 in response to cholesterol signaling. J. Biol. Chem. 280, 1901–1910.
Kathuria H., Cao Y.X., Ramirez M.I., Williams M.C. 2004. Transcription of the caveolin-1 gene is differentially regulated in lung type I epithelial and endothelial cell lines. A role for ETS proteins in epithelial cell expression. J. Biol. Chem. 279, 30028–30036.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © K.A. Arkhipova, I.B. Zborovskaya, 2012, published in Biologicheskie Membrany, 2012, Vol. 29, No. 6, pp. 387–399.
The article was translated by the authors.
Rights and permissions
About this article
Cite this article
Arkhipova, K.A., Zborovskaya, I.B. Microdomain-forming proteins of different families in common signal pathways. Biochem. Moscow Suppl. Ser. A 7, 1–11 (2013). https://doi.org/10.1134/S1990747812060037
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1990747812060037