Abstract
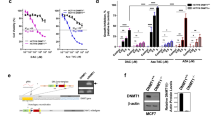
Cancer cells are characterized by hypermethylation of the promoter regions of tumor suppressor genes. DNA methyltransferase inhibitors reactivate the genes, pointing to DNA methyltransferases as potential targets for anticancer therapy. Dimeric bisbenzimidazoles varying in the length of an oligomeric linker between two bisbenzimidazole residues (DB(n), where n is the number of methylene groups in the linker) were earlier shown to efficiently inhibit methylation of DNA duplexes by murine DNA methyltransferase Dnmt3a. Here, some of the compounds were tested for cytotoxicity, cell penetration, and effect on genomic DNA methylation in F-977 fetal lung fibroblasts and HeLa cervical cancer cells. Within the 0–60 μM concentration range, only DB(11) exerted a significant toxic effect on normal cells, whereas the effects of DB(n) on cancer cells were not significant. DB(1) and DB(3) slightly stimulated proliferation of HeLa and F-977 cells, respectively. DB(1) and DB(3) penetrated into the nuclei of HeLa and F-977 cells and accumulated predominantly in or near the nucleolus, while DB(11) was incapable of nuclear penetration. HeLa cells incubated with 26 μM DB(1) or DB(3) displayed a decrease in methylation of the 18S rRNA gene, which was in the regions of predominant accumulation of DB(1) and DB(3). The same DB(3) concentration exerted a similar effect on F-977 cells. However, the overall genomic DNA methylation level remained unchanged in both of the cell lines. The results indicated that DB(n)-type compounds can be used to demethylate certain genes and are thereby promising as potential anticancer agents.
Similar content being viewed by others
Abbreviations
- DB(n):
-
dimeric bisbenzimidazole
- m5dC:
-
5-methyl-2′-deoxycytidine
- MTase:
-
C5-cytosine DNA methyl-transferase
- MTT:
-
3-[4,5-dimethylthyazol-2-yl]-2,5-diphe-nyltetrazolium bromide
- DMSO:
-
dimethyl sulfoxide
- PCR:
-
polymerase chain reaction
- rDNA:
-
ribosomal repeat
References
Bird A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21.
Hermann A., Gowher H., Jeltsch A. 2004. Biochemistry and biology of mammalian DNA methyltransferases. Cell Mol. Life Sci. 61, 2571–2587.
Eden A., Gaudet F., Waghmare A., Jaenisch R. 2003. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 300, 455.
Mani S., Herceg Z. 2010. DNA demethylating agents and epigenetic therapy of cancer. Adv. Genetics. 70, 327–340.
Kirsanova O.V., Cherepanova N.A., Gromova E.S. 2009. Inhibition of C5-cytosine-DNA methyltransferases. Biochemistry (Moscow). 74, 1175–1186.
Garcia-Manero G. 2008. Demethylating agents in myeloid malignancies. Curr. Opin. Oncol. 20, 705–710.
Streltsov S.A., Gromyko A.V., Oleinikov V.A., Zhuze A.L. 2006. The Hoechst 33258 covalent dimer covers a total turn of the double-stranded DNA. J. Biomol. Struct. Dynamics. 24, 285–302.
Cherepanova N.A., Ivanov A.A., Maltseva D.V., Minero A.S., Gromyko A.V., Streltsov S.A., Zhuze A.L., Gromova E.S. 2011. Dimeric bisbenzimidazoles inhibit the DNA methylation catalyzed by the murine Dnmt3a catalytic domain. J. Enzyme Inhibit. Med. Chem. 26, 295–300.
Veiko N.N., Liapunova N.A., Kosyakova N.V., Spitkovskii D.M. 2001. Elements of structural organization of the transcribed area of the ribosomal repeat (rDNA) in human peripheral blood lymphocytes. Mol. Biol. (Moscow). 35, 45–55.
Lyapunova N.A., Veiko N.N. 2010. Ribosomal genes in the human genome: Identification of four fractions, their organization in the nucleolus and metaphase chromosomes. Russ. J. Genet. 46, 1070–1073.
Costello J.F., Fruhwald M.C., Smiraglia D.J., Rush L.J., Robertson G.P., Gao X., Wright F.A., Fera-misco J.D., Peltomaki P., Lang J.C., Schuller D.E., Yu L., Bloomfield C.D., Caligiuri M.A., Yates A., Nishikawa R., Su Huang H., Petrelli N.J., Zhang X., O’Dorisio M.S., Held W.A., Cavenee W.K., Plass C. 2000. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nature Genet. 24, 132–138.
Esteller M. 2007. Epigenetic gene silencing in cancer: The DNA hypermethylome. Hum. Mol. Genet. 16(1), R50–R59.
Ren J., Singh B.N., Huang Q., Li Z., Gao Y., Mishra P., Hwa Y.L., Li J., Dowdy S.C., Jiang S.W. 2011. DNA hypermethylation as a chemotherapy target. Cell Signal. 23, 1082–1093.
Fang M.Z., Wang Y., Ai N., Hou Z., Sun Y., Lu H., Welsh W., Yang C.S. 2003. Tea polyphenol (-)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 63, 7563–7570.
Villar-Garea A., Fraga M.F., Espada J., Esteller M. 2003. Procaine is a DNA-demethylating agent with growth-inhibitory effects in human cancer cells. Cancer Res. 63, 4984–4989.
Lee B.H., Yegnasubramanian S., Lin X., Nelson W.G. 2005. Procainamide is a specific inhibitor of DNA methyltransferase 1. J. Biol. Chem. 280, 40749–40756.
Segura-Pacheco B., Trejo-Becerril C., Perez-Cardenas E., Taja-Chayeb L., Mariscal I., Chavez A., Acuna C., Salazar A.M., Lizano M., Duenas-Gonzalez A. 2003. Reactivation of tumor suppressor genes by the cardiovascular drugs hydralazine and procainamide and their potential use in cancer therapy. Clin. Cancer Res.: Official J. Am. Assoc. Cancer Res. 9, 1596–1603.
Datta J., Ghoshal K., Denny W.A., Gamage S.A., Brooke D.G., Phiasivongsa P., Redkar S., Jacob S.T. 2009. A new class of quinoline-based DNA hypomethylating agents reactivates tumor suppressor genes by blocking DNA methyltransferase 1 activity and inducing its degradation. Cancer Res. 69, 4277–4285.
Ivanov A.A., Salyanov V.I., Streltsov S.A., Cherepanova N.A., Gromova E.S., Zhuze A.L. 2011. DNA sequence-specific ligands: XIV. Synthesis of fluorescent biologicaly active dimeric bisbenzimidazoles, DB(3, 4, 5, 7, 11). Russ. J. Bioorg. Chem. 37, 472–482.
Petrova N.V., Iarovaia O.V., Verbovoy V.A., Razin S.V. 2005. Specific radial positions of centromeres of human chromosomes X, 1, and 19 remain unchanged in chromatin-depleted nuclei of primary human fibroblasts: Evidence for the organizing role of the nuclear matrix. J. Cell. Biochemistry. 96, 850–857.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © M.V. Darii, A.R. Rakhimova, V.N. Tashlitsky, S.V. Kostyuk, N.N. Veiko, A.A. Ivanov, A.L. Zhuze, E.S. Gromova, 2013, published in Molekulyarnaya Biologiya, 2013, Vol. 47, No. 2, pp. 292–301.
These authors contributed equally to this work.
Rights and permissions
About this article
Cite this article
Darii, M.V., Rakhimova, A.R., Tashlitsky, V.N. et al. Dimeric bisbenzimidazoles: Cytotoxicity and effects on DNA methylation in normal and cancer human cells. Mol Biol 47, 259–266 (2013). https://doi.org/10.1134/S0026893313020040
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893313020040