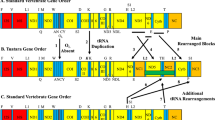
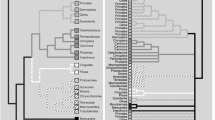
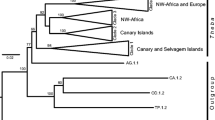
“...obtaining a strongly supported tree does not necessarily mean that it is correct.”
F. Delsuc, H. Brinkman, and H. Philippe, 2005
Abstract
The review considers the current problems of molecular phylogenetics based on mitochondrial and chromosomal DNA sequences. The emphasis is placed on mtDNA markers, which are widely employed in reconstructing molecular evolution, but often without a critical analysis of the physiological and biochemical features of mitochondria that affect the adequacy and reliability of the results. In addition to the factors that make mtDNA-based phylogenies difficult to interpret (unrecognized hybridization and introgression events, ancestral polymorphism, and nuclear paralogs of mtDNA sequences), attention is paid to the nonneutrality and unequal mutation rates of mtDNA genes and their fragments, violations of uniparental inheritance of mitochondria, recombination events, natural heteroplasmy, and mtDNA haplotypic diversity. These factors may influence the congruence of phylogenetic inferences and trees constructed for the same organisms with different mtDNA markers or with mitochondrial and nuclear markers. The review supports the viewpoint that mitochondrial genes and their fragments fail to provide reliable evolutionary markers when considered without a thorough study of the environmental conditions and life of the taxa. The influence of external conditions on the metabolism and physiology of mitochondria cannot be taken into account in full nor modeled well enough for phylogenetic applications. It is assumed that mtDNA is valuable as a phylogenetic marker primarily because its complete sequence may be analyzed to identify the apomorphic and synmorphic properties of a taxon and to search for informative nuclear paralogs of mtDNA for phylogeographical studies and estimations of relative evolution times.
Similar content being viewed by others
References
Moore W.S. 1995. Inferring phylogenies from mtDNA variation: Mitochondrial-gene trees versus nucleargene trees. Evolution. 49, 718–726.
Wiens J.J., Penkrot T.A. 2002. Delimiting species using DNA and morphological variation and discordant species limits in spiny lizards (Sceloporus). Syst. Biol. 51, 69–91.
Grechko V.V. Molecular markers in phylogeny and systematics. Russ. J. Genet. 38, 851–868.
Engstrom T.M., Schaffer H.B., McCord W.P. 2004. Multiple data set, high homoplasy, and the phylogeny of softshell turtles (Testudines: Trionychidae). Syst. Biol. 53, 693–710.
McCracken K.G., Sorenson M.D. 2005. Is homology or lineage sorting the source of incongruent mtDNA and nucler gene trees in the stiff-tailed ducks (Nomonycs-Oxyura)? Syst. Biol. 54, 35–55.
Skinner A., Donnellan S.C., Hutchinson M.N., Hutchinson R.G. 2005. A phylogenetic analysis of Pseudonaja (Hydrophiinae, Elapidae, Serpentes) based ob mitochondrial DNA sequences. Mol. Phyl. Evol. 37, 558–571.
Sorenson M.D., Quinn T.W. 1998. Numts: A challenge for avian systematics and population biology. The Auk. 115, 214–221.
Weisrock D.W., Smith S.D., Chan L.M., Biebouw K., Kappeler P.M., Yoder A.D. 2012. Concatenation and concordance in the reconstruction of the mouse lemur phylogeny: An empirical demonstration of the effect of allele sampling in phylogeny. Mol. Biol. Evol. doi 10.1093/molbev/mss008
Greaves S.N.J., Chapple D.G., Gleeson D.M., Daugherty C.H., Ritchie P.A. 2007. Phylogeography of the spotted skink (Oligosoma lineoocellatum) and green skink (O. chloronoton) species complex (Lacertilia: Scincidae) in New Zealand reveals pre-Pleistocene divergence. Mol. Phyl. Evol. 45, 729–739.
Solovyeva E.N., Poyarkov N.A., Dunaev E.A., Duysenbayeva T.N., Bannikova A.A. 2011. Molecular differentiation and taxonomy of the sunwatcher toadheaded agama species complex Phrynocephalus super-species helioscopus (Pallas, 1771) (Reptilia: Agamidae). Russ. J. Genet. 47, 842–856.
Brower A.V.Z., DeSalle R., Vogler A. 1996. Gene trees, species trees, and systematics: A cladistic perspective. Annu. Rev. Ecol. Syst. 27, 423–450.
Losos J.B., Jackman T.R., Larson A., De Queirroz K., Rodrigues-Shettino L. 1998. Contingency and determinism in replicated adaptive radiations of island lizards. Science. 279, 2115–2118.
Hanley T.C., Caccone A. 2005. Development of primers to characterization the mitochondrial control region of Galapagos land and marine iguanas (Conolophus and Ambleyrhynchus). Mol. Ecol. Notes. 5, 599–601.
Kurabayashi A., Sumida M., Yonekawa H., Glaw F., Hasegawa M. 2008. Phylogeny, recombination, and mechanisms of stepwise mitochondrial genome reorganization in mantelliid frogs from Madagascar. Mol. Biol. Evol. 25, 874–891.
Ballard J.W.O., Whitlock M.C. 2004. The incomplete natural history of mitochondria. Mol. Ecol. 13, 729–744.
Moritz C., Brown W.M. 1987. Tandem duplications in animal mitochondrial DNAs: Variation in incidence and gene content among lizards. Proc. Natl. Acad. Sci. U. S. A. 84, 7183–7187.
Brown W.M., Prager E.M., Wang A., Wilson A.C. 1982. Mitochondrial DNA sequences of primates: Tempo and mode of evolution. J. Mol. Evol. 18, 225–239.
Tsaousis A.D., Martin D.P., Ladoukakis E.D., Posada D., Zouros E. 2005. Widespread recombination in published animal mtDNA sequences. Mol. Biol. Evol. 22, 925–933.
Castellana S., Vicario S., Saccone C. 2011. Evolutionary patterns of the mitochondrial genome in Metazoa: Exploring the role of mutation and selection in mitochondrial protein-coding genes. Genome Biol. Evol. 3, 1067–1079.
Moritz C., Dowling T.E., Brown W.M. 1987. Evolution of the animal mitochondrial DNA: Relevance for population biology and systematics. Annu. Rev. Ecol. Syst. 18, 269–292.
Crother B.I., Presh W. 1994. Xantusiid lizards, concern for analysis, and the search for a best estimate of phylogeny: Furter comments. Mol. Phyl. Evol. 3, 272–275.
Hedges S.B., Bazy R.L. 1994. Reply: Xantusiid lizards and phylogenetic inference. Mol. Phyl. Evol. 3, 275–278.
Palumbi S.R., Baker C.S. 1994. Contrasting population structure from nuclear intron sequences and mtDNA of humpback whales. Mol. Biol. Evol. 11, 426–435.
Ballard J.W., Kreitman M. 1995. Is mitochondrial DNA a strictly neutral marker? Trends Ecol. Evol. 10, 485–489.
Lunt D.H., Hymen B.C. 1997. Animal mitochondrial DNA recombination. Nature. 387, 247.
Curole J.P., Kocher T.D. 1999. Mitogenomics: Digging deeper with complete mitochondrial genomes. Trends Ecol. Evol. 14, 394–308.
Vorontsov N.N. 1999. Razvitie evolyutsionnykh idei v biologii (Development of Evolution Ideas in Biology). Moscow: ABF Publ.
Ballard J.W.O., Chernoff B., James A.C. 2002. Divergence of mitochondrial DNA is not corroborated by nuclear DNA, morphology, or behaviour in Drosophila simulans. Evolution. 56, 527–545.
Zink R.M., Barrowclough G.F. 2008. Mitochondrial DNA under siege in avian phylogeography. Mol. Ecol. 17, 2107–2121.
Shoo L.P., Rose R., Doughty P., Austin J.J., Melville J. 2008. Diversification patterns of pebble-mimic dragons are consistent with historical disruption of important habitat corridors in arid Australia. Mol. Phyl. Evol. 48, 528–542.
Edwards S.V. 2009. Is a new and general theory of molecular systematics emerging? Evolution. 63, 1–19.
Camargo A., Sinervo B., Sites J.W., jr. 2010. Lizards as a model organisms for linking phylogeographic and speciation studies. Mol. Ecol. 19, 3250–3270.
Planet P.J. 2006. Tree disagreement: Measuring and testic incongruence in phylogenies. J. Biomed. Inform. 39, 86–102.
Verdue-Ricoy J., Carranza S., Salvator A., BU. S. Ack S.D., Diaz J.A. 2010. Phylogeography of Psammodromus algirus (Lacertidae) revisited: Systematic implications. Amphibia-Reptilia. 31, 576–582.
Godinho R., Crespo E.G., Ferrand N. 2008. The limits of mtDNA phylogeography: Complex patterns of population history in the highly structured Iberian lizard are only revealed by the use of nuclear markers. Mol. Ecol. 17, 4670–4683.
Dolman G., Moritz C. 2006. A multilocus perspective on refugial isolation and divergence in reinforest skinks (Carlia). Evolution. 60, 573–582.
Sequiera F., Alexandrino J., Wess S., Ferrand N. 2008. Documenting the advantage and limitations of different classes of molecular markers in a well-established phylogeographic context: Lessons from the Iberian endemic golden-stripped salamander, Chioglossa lusitanica (Caudata: Salamandridae). Biol. J. Linn. Soc. 95, 371–387.
Leache A.D. 2010. Species tree for spiny lizards (genus Sceloporus): Identifying points of concordance and conflict between nuclear and mitochondrial data. Mol. Phyl. Evol. 54, 162–171.
Fisher-Reid M.C., Wiens J.J. 2011. What are the consequences of combining nuclear sand mitochondrial data for phylogenetic analysis? Lessons from Plethodon salamanders and 13 other vertebrate clades. BMC Evol. Biol. 11, 300–320.
Korpelainen H. 2004. The evolutionary processes of mitochondrial and chloroplast genomes differ from those of nuclear genomes. Naturwissenschaften. 91, 505–518.
Degnan J.H., Rosenberg N.A. 2006. Discordance of species trees with their most likely gene trees. PLoS Genet. 2, e68.
Holland B.R., Benthin S., Lockart P.J., Moulton V., Huber K.T. 2008. Using supernetworks to distinguish hybridization from lineage-sorting. BMC Evol. Biol. 8, 202–213.
Degnan J.H., Rosenberg N.A. 2009. Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol. Evol. 24, 332–340.
Hickerson M.J., Carstens B.C., Cavender-Bares J., Crandall K.A., Graham C.H., Johnson J.B., Rissler L., Victoriano P.F., Yoder A.D. 2010. Phylogeography’s past, present, and future: 10 years after Avise, 2000. Mol. Phyl. Evol. 54, 291–301
Pollard D.A., Iyer V.N., Moses A.M., Eisen M.B. 2006. Widespread discordance of gene trees with species tree in Drosophila: Evidence for incomplete lineage sorting. PLoS Genet. 2, e173.
MacGuire J.A., Linkem C.W., Koo M.S., Hutchinson D.W., Lappin A.K., Orange D.I., Lemos-Espinal J., Riddle B.R., Jaeger J.R. 2007. Mitochondrial introgression and incomplete lineage sorting through space and time: Phylogenetics of crotaphytid lizards. Evolution. 61, 2879–2897.
Knowless L.L., Carstens B.C. 2007. Delimiting species without monophyletic gene trees. Syst. Biol. 56, 887–895.
Leache A.D., Koo M.S., Spencer C.L., Papenfuss T.J., Fischer R.N. 2009. Quantifying ecological, morphological, and genetic variation to delimit species in the coast horned lizard species complex (Phrynosoma). Proc. Natl. Acad. Sci. U. S. A. 106, 12418–12423.
Benavides E., Baum R., McClellan D., Sites J.W., jr. 2007. Molecular phylogenetics of the lizard genus Microlophus (Squamata: Tropiduridae): Aligning and retrieving indel signals from nuclear introns. Syst. Biol. 56, 776–797.
Kmiec B., Woloszynska M., Janska H. 2006. Heteroplasmy as a common state of mitochondrial genetic information in plants and animals. Curr. Genet. 50, 149–159.
Grechko V.V. 2011. Repeated DNA sequences as an engine of biological diversification. Mol. Biol. (Moscow) 45, 704–727.
Leache A.D., McGuire J.A. 2006. Phylogenetic relationships of horned lizards (Phrynosoma) based on nuclear and mitochondrial data: Evidence for a misleading mitochondrial gene tree. Mol. Phyl. Evol. 39, 628–644.
Leache A.D., Reeder T.W. 2002. Molecular systematics of the eastern fence lizard (Sceloporus undulates): A comparison of parsimony, likelihood, and Bayesian approaches. Syst. Biol. 51, 44–68.
Douglas D.A., Arnason U. 2009. Examining the utility of categorical models and alleviating artifacts in phylogenetic reconstruction of the Squamata (Reptilia). Mol. Phyl. Evol. 52, 784–796.
Lindell J., Mendez-de la Cruz. F.R., Murphy R.W. 2005. Deep genealogical history without population differentiation: Discordance between mtDNA and allozyme divergence in the zebra-tailed lizard (Gallisaurus draconoides). Mol. Phyl. Evol. 36, 682–694.
Rowlings L.H., Rabosky D.L., Donnellan S.C., Hutchinson M.N. 2008. Python phylogenetics: Inference from morphology and mitochondrial DNA. Biol. J. Linn. Soc. 93, 603–619.
Honda M., Ota H., Murphy R.W., Hikida T. 2006. Phylogeny and biogeography of water skinks of the genus Tropidophorus (Reptilia: Scincidae): A molecular approach. Zool. Scripta. 35, 85–95.
Yang Z., Rannala B. 2010. Bayesian species delimitation using multilocus sequence data. Proc. Natl. Acad. Sci. U. S. A. 107, 9264–9269.
Russo C.A.M., Takezaki N., Nei M. 1996. Efficiency of different genes and the different tree-building methods in recovering a known vertebrate phylogeny. Mol Biol. Evol. 13, 525–536.
Giribet G., Wheeler W.C. 1999. On gaps. Mol. Phyl. Evol. 13, 132–143.
Simmons M.P., Ochoterena H. 2000. Gaps as character in sequence-based phylogenetic analysis. Syst. Biol. 49, 369–381.
Miclosh I., Lunter G.A., Holmes I. 2004. A “long indel” model for evolutionary sequence alignment. Mol. Biol. Evol. 21, 529–540.
Ashton K.G., De Queiroz A. 2001. Molecular systematics of the western rattlesnake, Crotalus viridis (Viperidae), with comments on the utility of the Dloop in the phylogenetic studies of snakes. Mol. Phyl. Evol. 21, 176–189.
Gatesy J., Baker R.H. 2005. Hidden likelihood support in genomics data: Can forty-five wrongs make a right? Syst Biol. 54, 483–492.
Gadagkar S.R., Rosenberg M.S., Kumar S. 2005. Inferring species phylogenies from multiple genes: Concatenated sequence tree versus consensus gene tree. J. Exp. Zool. (Mol. Dev. Evol.) 304B, 64–74.
Kubatko L.S., Degnan J.H. 2007. Inconsistency of phylogenetic estimates from concatenated data under coalescence. Syst. Biol. 56, 17–24.
Liu L., Pearl D.K., Brumfield R.T., Edwards S.V. 2008. Estimating species trees ueing multiple-allele DNA sequence data. Evolution. 62, 2080–2091.
Maddison W.P., Knowless L.L. 2006. Inferring phylogeny despite incomplete lineage sorting. Syst. Biol. 55, 21–30.
Keogh I.S., Edwards D.L., Fisher R.N., Harlow P.S. 2008. Molecular and morphological analysis of the critically endangered Fijian iguanas reveals cryptic diversity and a complex biogeographic history. Phil. Trans. R. Soc. B. doi:10.1098/rstb.2008.0120
Wiens J.J., Hollingsworth B.D. 2000. War of the iguanas: Conflicting molecular and morphological phylogenies and long-branch attraction in iguanid lizards. Syst. Biol. 49, 143–159.
Oliviero M., Bolgna M.A., Mariottini P. 2000. Molecular biogeography of the Mediterranean lizards Podarcis Wagler, 1830, and Teira Gray, 1838 (Reptilia, Lacertidae). J. Biogeogr. 27, 1403–1420.
Slowinsky J.B. 2001. Molecular polytomies. Mol. Phyl. Evol. 18, 114–120.
Suzuki Y., Glazko G.V., Nei M. 2002. Overcredibility of molecular phylogenies obtained by Bayesian phylogenetics. Proc. Natl. Acad. Sci. U. S. A. 99, 16138–16143.
Townsend T., Larson A. 2002. Molecular phylogenetics and mitochondrial genomic evolution in the Chamaeleonidae (Reptilia, Squamata). Mol. Phyl. Evol. 23, 22–36.
Jennings W.B., Pianka E.R., Donnellan S. 2003. Systematics of the lizard family Pygopodidae with implications for the diversification of Australian temperate biota. Syst. Biol. 52, 757–780.
Kolaczkowski B., Thornton J.W. 2004. Performance of maximum parsimony and likelihood phylogenetics when evolution is heterogeneous. Nature. 431, 980–984.
Castoe T.A., Parkinson C.L. 2006. Bayesian mixed models and the phylogeny of pitvipers (Viperidae: Serpentes). Mol. Phyl. Evol. 39, 91–110.
Melvill J., Ritchie E.G., Chapple S.N.J., Glor R.E., Schulte J.A. II. 2011. Evolutionary origins and diversification of dragon lizards in Australia’s tropical savannas. Mol. Phyl. Evol. 58, 257–270.
Petit R.J., Excoffier L. 2009. Gene flow and species delimitation. Trends Ecol. Evol. 24, 386–395.
Zamudio K.R., Jones K.B., Ward R.H. 1997. Molecular systematics of short-horned lizards: Biogeography and taxonomy of a widespread species complex. Syst. Biol. 46, 284–305.
Pang J., Wang Y., Zhong Y., Hoelzerl A.R., Papenfuss T.J., Zeng X., Ananjeva N.B., Zhang Y. 2003. A phylogeny of Chinese species in the genus Phrynocephalus (Agamidae) inferred from mitochondrial DNA sequences. Mol. Phyl. Evol. 27, 398–409.
Melvill J., Hale J., Mantziou G., Ananjeva N.B., Milto K. 2009. Historical biogeography, phylogenetic relationships and intraspecific diversity of agamid lizards in the Central Asian deserts of Kazakhstan and Uzbekistan. Mol. Phyl. Evol. 53, 99–112.
Castoe T.A, De Koning A.P.J, Kim H.-M., Gu W., Noonan B.P., Naylor G., Jiang Z.J., Parkinson C.L., Pollock D.D. 2009. Evidence for an ancient adaptive episode of convergent molecular evolution. Proc. Natl. Acad. Sci. U. S. A. 106, 8986–8991.
Templeton A.R. 2009. Why does a method that fails continue to be used? The answer. Evolution. 63, 807–812.
Templeton A.R. 2010. Coalescent-based, maximum likelihood inference in phylogeography. Mol. Ecol. 19, 431–435.
Knowless L.L. 2008. Why does a method that fails continue to be used? Evolution. 62, 2713–2717.
Beaumont M.A., Nielsen R., Robert C., et al. 2010. In defence of model-based inference in phylogeography. Mol. Ecol. 19, 436–446.
Paulo O.S., Jordan W.C., Bruford M.W., Nichols R.A. 2002. Using nested clade analysis to assess the history of colonization and the persistence of populations of the Iberian lizard. Mol. Ecol. 11, 809–819.
Morando M., Avila L.J., Sites J.W., jr. 2003. Sampling strategies for delimiting species: genes, individuals, and populations in the Liolaemus elongates-kriegi complex (Squamata: Liolaemidae) in Andean-Patagonian South America. Syst. Biol. 52, 159–185.
Gifford M.E., Larson A. 2008. In situ genetic difference in Ameiva chrysolaemus: Multilocus perspective. Mol. Phyl. Evol. 49, 277–291
O’Meara B.C. 2010. New heuristic methods for joint species delimitation and species tree inference. Syst. Biol. 59, 59–73.
Chapple D.G., Keogh J.S., Hutchinson M.N. 2004. Molecular phylogeography and systematics of the arid-zone members of the Egernia whitii (Lacertilia: Scincidae) species group. Mol. Phyl. Evol. 33, 549–561.
Giribet G., DeSalle R., Wheeler W.C. 2002. “Pluralism” and the aims of phylogenetic research. In: Molecular Systematics and Evolution. Basel: Birkhauser Verlag, p. 141.
Eckert A.J., Carstens B.C. 2008. Does gene flow destroy phylogenetic signal? The performance of three methods for estimating species phylogenies in the presence of gene flow. Mol. Phyl. Evol. 49, 832–842.
Heled J., Drummond A.J. 2010. Bayesian inference of species trees from multilocus data. Mol. Biol. Evol. 27, 570–580.
Delsuc F., Brinkmann H., Philippe H. 2005. Phylogenomics and the reconstruction of the Tree of Life. Nature Rev. Genet. 6, 361–375.
Alifanov V.R. 2007. Lizards in the dinosaurus era. Priroda (Moscow) 9, 47–58.
Janes D. E., Organ C.L., Fujita M.K., Shedlock A.M., Edwards S.V. 2010. Genome evolution in Reptilia, the sister group of Mammals. Annu. Rev. Genom. Hum. Genet. 11, 239–264.
Hedges S.B., Vidal N. 2009. Lizards, snakes, and amphisbaenians (Squamata). In: The Timetree of Life. Eds. Hedges S.B., Kumar S. Oxford: Oxford Univ. Press, pp. 383–389.
Sites J.W., Davis S.K., Guerra T., Iverson J.B., Snell H.L. 1996. Character congruence and phylogenetic signal in molecular and morphological data sets: A case study in the living iguanas (Squamata, Iguanidae). Mol. Biol Evol. 13, 1087–1105.
Macey J.R., Larson A., Ananjeva N.B., Papenfuss T.J. 1997. Evolutionary shifts in the three major structural features of the mitochondrial genome among iguanian. J. Mol. Biol. 44, 660–674.
Fuller S., Baverstock P., King D. 1998. Biogeographic origins of goannas (Varanidae): A molecular perspective. Mol. Phyl. Evol. 9, 294–307.
Harris D.J. 1999. Molecular systematics and evolution of lacertid lizards. Nat. Croat. 8, 161–180.
Frost D.R., Rodrigues M.T., Grant T., Titus T.A. 2001. Phylogenetics of the lizards genus Tropidurus (Squamata: Tropiduridae: Tropidurinae): Direct optimization, descriptive efficiency, and sensitivity analysis of congruence between molecular data and morphology. Mol. Phyl. Evol. 21, 352–371.
Bromham L., Woolfit M., Lee M.S.Y., Rambaut A. 2002. Testing the relationship between morphological and molecular rates of change along phylogenies. Evolution. 56, 1921–1930.
Schulte J.A. II, Valladares J.P., Larson A. 2003. Phylogenetic relationships within Iguanidae inferred using molecular and morphological data and a phylogenetic taxonomy of iguanian lizards. Herpetologica. 56, 399–319.
Whiting A.S., Bauer A.M., Sites J.W., jr. 2003. Phylogenetic relationships and limb loss in sub-Saharan African scincine lizards (Squamata: Scincidae). Mol. Phyl. Evol. 29, 582–598.
Townsend T.M., Larson A., Louis E., Macey J.R. 2004. Molecular phylogenetics of Squamata: The position of snakes, amphisbaenians, and dibamids, and the root of the squamate tree. Syst. Biol. 53, 735–757.
Vidal N., Hedges S.B. 2009. The molecular evolution tree of lizards, snakes and amphysbaenians. C. R. Biologies. 332, 129–139.
Carranza S., Arnold E.N. 2006. Systematics, biogeography, and evolution of Hemidactylus geckos (Reptilia: Gekkonidae) elucidated using mitochondrial DNA sequences. Mol. Phyl. Evol. 38, 531–545.
Fry B.G., Vidal N., Norman J.A., Vonk F.J., Scheib H., Ramjan S.F.R., Kuruppu S., Fung K., Hedges S.B., Richardson M.K., Hodgson W.C., Ignjatovic V., Summerhayes R., Kochva E. 2006. Early evolution of the venom system in lizards and snakes. Nature. 439, 584–588.
Arnold E.N., Arribas O., Carranza S. 2007. Systematics of the Palaearctic and Oriental lizard tribe Lacertini (Squamata: Lacertidae: Lacertinae), with description of eight new genera. Zootaxa. 1430, 1–86.
Kumazawa Y. 2007. Mitochondrial genome from major lizard families suggest their phylogenetic relationships and ancient radiation. Gene. 388, 19–26.
Carretero M.A. 2008. An integrated assessment of a group with complex systematics: Iberomaghrebian lizard genus Podarcis (Squamata: Lacertidae). Integr. Zool. 4, 247–266.
Kaply P., Limberakis P., Poulakakis N., Mantziou G., Parmakelis A., Mylonas M. 2008. Molecular phylogeny of three Mesalina (Reptilia: Lacertidae) species (M. gattulata, M. brevirostris, and M. bahaeldini) from the North Africa and the Middle East: Another case of paraphyly? Mol. Phyl. Evol. 49, 102–110.
Conrad J.L. 2008. Phylogeny and systematics of Squamata (Reptilia) based on morphology. Bull. Am. Mus. Nat. Hist. 310, 1–182.
Keogh J.S. 1998. Molecular phylogeny of elapid snakes and a consideration of their biogeographic history. Biol. J. Linn. Soc. 63, 177–203.
Dong S., Kumazawa Y. 2005. Complete mitochondrial DNA sequences of six snakes: Phylogenetic relationships and molecular evolution of genomic features. J. Mol. Evol. 61, 12–22.
Lee M.S., Hugall A.F., Lawson R., Scanlon J.D. 2007. Phylogeny of snakes (Serpentes): Combining morphological and molecular data in likelihood, Bayesian and parsimony analyses. Syst. Biodivers. 5, 371–389.
Voris H.K., Karns D.R., Feldheim K.A., Kechavarti B., Rinehart M. 2008. Multiple paternity in the Oriental-Australian rear-ranged watersnake (Homalopsidae). Herp. Cons. Biol. 3, 88–102.
Alfaro M.E., Karns D.R., Voris H.K., Brock C.D., Stuart B.L. 2008. Phylogeny, evolutionary history, and biogeograpny of Oriental-Australian rear-ranged water snakes (Colubroidea: Homalopsidae) inferred from mitochondrial and nuclear DNA sequences. Mol. Phyl. Evol. 46, 576–593.
Rest J.S., Ast J.C., Austin C.C., Waddell P.J., Tibbetts E.A., Hay J.M., Mindell D.P. Molecular systematics of primary reptilian lineages and the tuatara mitochondrial genome. Mol. Phyl. Evol. 29, 289–297.
Bryson R.W. Jr., Pastorini J., Burbrink F.T., Forstner M.R.J. 2007. A phylogeny of the Lampropeltis mexicana (Serpentes: Colubridae) based on mitochondrial DNA sequences suggests evidence for species-level polyphyly within Lampropeltis. Mol. Phyl. Evol. 43, 674–684.
Cox S.C., Carranza S., Brown R.P. 2010. Divergence times and colonization of the Canary Islands by Gallotia lizards. Mol. Phyl. Evol. 56, 747–757.
Guicking D., Lawson R., Joger U., Wink M. 2006. Evolution and phylogeny of the genus Natrix (Serpentes: Colubridae). Biol. J. Linn. Soc. 87, 127–143.
Nardi F., Carapelli A., Fanciully P.P., Dallai R., Frati F. 2001. The complet mitochondrial DNA sequence of the basal hexapod Tetrodontophora bielanensis: Evidence for heteroplasmy and tRNA translocations. Mol. Biol. Evol. 18, 1923–1304.
Lawson R., Slowinski J.B., Crother B.I., Burbink F.T. 2005. Phylogeny of the Colubroidea (Serpentes): New evidence from mitochondrial and nuclear genes. Mol. Phyl. Evol. 37, 581–601.
Torres-Carvajal O., Schulte J.A. II, Cadle J.E. 2006. Phylogenetic relationships of South American lizards of the genus Stenocercus (Squamata: Iguania): A new approach using a general mixture model for gene sequence data. Mol. Phyl. Evol. 39, 171–185.
Moritz C. 1994. Defining “evolutionary significant units” for cobservation. Trends Ecol. Evol. 9, 373–375.
Honda M., Ota H., Sengoku S., Yong H.-S., Hikida T. 2002. Molecular evaluation of phylogenetic significances in the highly divergent karyotypes of the genus Gonocephalus (Reptilia: Agamidae) from tropical Asia. Zool. Sci. 19, 129–133.
Gillooly J.F., Allen A.P., West G.B., Brown J.H. 2005. The rate of DNA evolution: Effects of body size and temperature on the molecular clock. Proc. Natl. Acad. Sci. U. S. A. 102, 140–145.
Rand D.M. 1994. Thermal habit, metabolic rate and the evolution of mitochondrial DNA. Trends Ecol. Evol. 9, 125–131.
Birky C.W., jr. 2001. The inheritance of genes in mitochondria and chloroplasts: Laws, mechanisms, and models. Annu. Rev. Genet. 35, 125–148.
Nunn G.B., Stanley S.E. 1998. Body size effects and rates of cytochrome c evolution in tube-nosed seabirds. Mol. Biol. Evol. 15, 1360–1371.
Soudheimer N., Glatz C.E., Tirone J.E., Deardorf M.A., Krieger A.M., Hakonarson H. 2011. Neutral mitochondrial heteroplasmy and the influence of aging. Hum. Mol. Genet. 20, 1653–1659.
Bromham L. 2002. Molecular clocks in reptiles: Life history influences the rate of molecular evolution. Mol. Biol. Evol. 19, 302–309.
Vitt L.J., Pianka E.R. 2004. Historical patterns in lizard ecology: What teiids can tell us about lacertids. In: The Biology of Lacertid Lizards. Evolutionary and Ecological Perspectives. Eds. Perrez-Melado V., Riera V., Perere A. Institut Menorqui d’Estudis, vol. 8, pp. 139–157.
Droge W. 2002. Free radicals in the physiological control of cell function. Physiol. Rev. 82, 47–95.
Wallace D.C. 2010. Mitochondrial DNA mutations in desease and aging. Environ. Mol. Mutagen. 51, 440–450.
Daniels S.R., Heideman N.J.L., Hendrics M.G.J., Mokone M.E., Crandall K.A. 2005. Unravelling evolutionary lineages in the limbless fossorial skinks genus Acontias (Sauria: Scincidae): Are subspecies equivalent systematic units? Mol. Phyl. Evol. 34, 645–654.
Edwards D.L., Melvill J. 2011. Extensive phylogeographic and morphological diversity in Diporiphore nobbi (Agamidae) leads to a taxonomic review and a new species description. J. Herpetol. 45, 530–546.
Cinnery P.F., Dahl H.H.M. 2000. The inheritance of mitochondrial DNA heteroplasmy: Random drift, selection, or both? Trends Genet. 16, 500–505.
Zhao X., Li N., Guo W., Hu X., Liu Z., Gong G., Wang A., Feng J., Wu C. 2004. Further evidence for paternal inheritance of mtDNA in the sheep (Ovis aries). Heredity. 93, 399–403.
Schwartz M., Vissing J. 2002. Paternal inheritance of mitochondrial DNA. N. Engl. J. Med. 347, 576–580.
Kraytsberg Y., Schwartz M., Brown T.A., Ebralidse K., Kunz W.S., Clayton D.A., Vissing J., Khrapko K. 2004. Recombination of human mitochondrial DNA. Science. 304, 981.
Laloi D., Richard M., Lecomte J., Massot L., Clobert J. 2004. Multiple paternity in clutches of common lizard Lacerta vivipara: Data from microsatellite markers. Mol. Ecol. 13, 719–723.
Grzybowski T., Malyarchuk B.A., Czarny J., Miscicka-Sliwka., Kotzbach R. 2003. High level of mitochondrial DNA heteroplasmy in single hair roots: Reanalysis and revision. Electrophoresis. 24, 1159–1165.
Wallis G.P. 1999. Do animal mitochondrial genomes recombine? Trends Ecol. Evol. 14, 209–210.
Gyllensten U.B., Wharton D., Joseffson A., Wilson A.C. 1991. Paternal inheritance of mitochondrial DNA in mice. Nature. 352, 255–257.
Hagelberg E. 2003. Recombination or mutational rates heterogeneity? Implications for mitochondrial Eve. Trends Genet. 19, 84–90.
Podnar M., Meyer W., Tvrtkovic N. 2005. Biogeography of the Italian wall lizard, Podarcis sicula, as revealed by mitochondrial DNA sequences. Mol. Ecol. 14, 575–588.
Townsend T.M., Larson A., Louis E., Macey J.R. 2004. Molecular phylogenetics of Squamata: The position of snakes, amphysbaenians, and dibamids, and the root of the Squamata. Syst. Biol. 53, 735–757.
Croucher P.J.P., Oxford G.S., Searle J.B. 2004. Mitochondrial differentiation, introgression and phylogeny of species in the Tegenaria atrica group (Araneae: Agelenidae). Biol. J. Linn. Soc. 81, 79–89.
Densmore L.D., Wright J.W., Brown W.M. 1985. Length variation and heteroplasmy are frequent in mitochondrial DNA from parthenogenetic and bisexual lizards (genus Cnemidophorus). Genetics. 110, 689–707.
Fonseca M.M., Brito J.C., Paulo O.S., Carretero M.A., Harris D.J. 2009. Systematic and phylogeographic assessment of the Acanthodactilus erythrurus group (Reptilia: Lacertidae) based on phylogenetic analyses of mitochondrial and nuclear DNA. Mol. Phyl. Evol. 51, 131–142.
Jenuth J.P., Peterson A.C., Shoubridge E.A. 1997. Tissue-specific selection for different mtDNA genotypes in heteroplasmic mice. Nature Genet. 16, 93–95.
Ast J.C. 2001. Mitochondrial DNA evidence and evolution in Varanoidea (Squamata). Cladistics. 17, 211–226.
Petri B., von Haeseler A., Paabo S. 1996. Extreme sequence heteroplasmy in bat mitochondrial DNA. Biol. Chem. 377, 661–667.
Frey J.E., Frey B. 2004. Origin of intra-individual variation in PCR-amplified mitochondrial cytochrome oxidase I of Thrips tabaci (Thysanoptera: Thripidae): Mitochondrial heteroplasmy or nuclear integration? Hereditas. 140, 92–98.
Funk D.J., Omland K.E. 2003. Species-level paraphyly: Frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annu. Rev. Ecol. Evol. Syst. 34, 397–423.
Reiner J.E., Kishare R.B., Levin B.C., Albanetti T., Boire N., Knipe A., Helmerson K., Deckman K.H. 2010. Detection of heteroplasmic mitochondrial DNA in single mitochondria. PLoS ONE. 5, e14359.
Nachman M.W., Brown W.M., Stoneking M., Aquadro C.F. 1996. Nonneutral mitochondrial DNA variation in humans and chimpanzees. Genetics. 142, 953–963.
Jackman T.R., Irschnic D.J., De Quairroz K., Losos J.B., Larson A. 2002. Molecular phylogenetic perspective on evolution of lizards of the Anolis grahami series. J. Exp. Zool. (Mol. Biol. Dev. Evol.). 294B, 1–16.
Thorpe R.S., Leadbeater D.L., Pook C.E. 2005. Molecular clock and geological dates: Cytochrome b of Anolis extremus substantially contradicts dating of Barnados emergence. Mol. Ecol. 14, 2087–2096.
Pereira S.L., Baker A.J. 2006. A multigenomic timescale for birds defects variable phylogenetic rates of molecular evolution and refute the standart molecular clock. Mol. Biol. Evol. 23, 1731–1740.
Smith S.A., Sadler R.A., Bauer A.M., Austin C.C., Jackman T. 2007. Molecular phylogeny of the scincid lizards of New Caledonia and adjacent areas: Evidence for a single origin of the endemic skinks of Tasmania. Mol. Phyl. Evol. 43, 1151–1166.
Weinreich D.M., Rand D.M. 2000. Contrasting patterns of nonneutral evolution in proteins encoded in nuclear and mitochondrial genomes. Genetics. 156, 385–399.
Bazin E., Glemin S., Galtier N. 2006. Population size does not influence mitochondrial genetic diversity in animals. Science. 312, 570–572.
Hudson R.R., Torelli M. 2003. Stochasticity overrules the “three-times rule”: Genetic drift, genetic draft, and coalescence times for nuclear loci versus mitochondrial DNA. Evolution. 57, 182–190.
Irwin D.M., Kocher T.D., Wilson A.C. 1991. Evolution of the cytochrome b gene of mammals. J. Mol. Evol. 32, 128–144.
Lopez P., Casane D., Philippe H. 2002. Heterotachy, an important process of protein evolution. Mol. Biol. Evol. 19, 1–7.
Jiang Z.J., Castoe T.A., Austin C.C., Burbrink F.T., Herron M.D., McGuire J.A., Parkinson C.L., Pollock D.D. 2007. Comparative mitochondrial genomics of snakes: Extraordinary substitution rate dynamics and functionality of the duplicated control region. BMC Evol. Biol. 7, 123–137.
Kumazawa Y. 2004. Mitochondrial DNA sequences of five squamates: Phylogenetic affiliation of snakes. DNA Res. 11, 137–144.
Hebert P.D.N., Cywinska A., Ball S.L., De Waard J.R. 2003. Bilogical identifications through DNA barcodes. Proc. R. Soc. London. 270, 313–321.
Norman J.E., Gray M.W. 2001. A complex organization of the gene encoding cytochrome oxidase subunit 1 in the mitochondrial genome of the dinoflagellate, Crypthecodinium cohnii: Homologous recombination generates two different cox1 open reading frames. J. Mol. Evol. 53, 351–363.
Rubinoff D., Cameron S., Will K. 2006. A genomic perspective on the shortcomings of mitochondrial DNA for “barcoding” identification. J. Hered. 97, 581–594.
Harris D.J., Sa-Sousa P. 2001. Species distinction and relationships of the western Iberian Podarcis lizards (Reptilian, Lacertidae) based on morphology and mitochondrial DNA sequences. Herpetol. J. 11, 129–136.
Meyer A. 1994. Shortcomings of the cytochrome b gene as molecular marker. Trends Ecol. Evol. 9, 278–280.
Kocher T.D., Thomas W.K., Meyer A., Edwards S.V., Paabo S., Villablanca F.X., Wilson A.C. 1989. Dynamics of mitochondrial DNA evolution in animals: Amplification and sequencing with conserved primers. Proc. Natl. Acad. Sci. U. S. A. 86, 6196–6200.
Castoe T., De Koning A.P., Kim H.-M., Gu W., Noonan B.P., Naylor G., Jiang Z.J., Parkinson C.L., Pollock D.D. 2009. Evidence for an ancient adaptive episode of convergent molecular evolution. Proc. Natl. Acad. Sci. U. S. A. 106, 8986–8991.
Albert E.M., San Mauro D., Garcia-Paris M., Ruber L., Zardoya R. 2009. Effect of taxon sampling on recovering the phylogeny of squamate reptiles based on complete mitochondrial genome and nuclear gene sequence data. Gene. 441, 12–21.
Baker R.J., Bradly R.D. 2006. Speciation in mammals and the genetic species concept. J. Mammal. 87, 643–662.
Chen Q.-L., Tang X.-S., Yao W.-J., Lu S.-Q. 2009. Bioinformatics analysis the complete sequences of cytochrome b of Takydromus sylvaticus and modeling the tertiary structure of encoded protein. Int. J. Biol. Sci. 5, 596–602.
Rokas A., Ladoukakis E., Zouros E. 2003. Animal mitochondrial DNA recombination revisited. Trends Ecol. Evol. 18, 411–417.
Shierup M.H., Hein J. 2000. Consequences of recombination on traditional phylogenetic analysis. Genetics. 156, 879–891.
Posada D., Crandal K.A. 2002. The effect of recombination on the accuracy of phylogenetic estimation. J. Mol. Evol. 54, 396–402.
Piganeau G., Gardner M., Eyre-Walker A. 2004. A broad survey of recombination in animal mitochondria. Mol. Biol. Evol. 21, 2319–2325.
Macey J.R., Schulte J.A., Ananjeva N.B., Larson A., Rastegar-Pouyani N., Shammakov S.M., Pappenfus T.J. 1998. Phylogenetic relationships among agamid lizards of Laudakia caucasica species group: Testing hypothesis of biogeographical fragmentation and an area cladogram for the Iranian plateau. Mol. Phyl. Evol. 10, 118–131.
Hoarau G., Holla S., Lescasse R., Stam W.T., Olsen J.L. 2002. Heteroplasmy and evidence for recombination in the mitochondrial control region of the flatfish Plathichtys felesus. Mol. Biol. Evol. 19, 2261–2264.
Shao R., Mitani H., Barker S.C., Takahasi M., Fukunaga M. 2005. Novel mt gene content and gene arrangement indicate illegitimate inter-mtDNA recombination in the chigger mite, Leptotrombidium pallidum. J. Mol. Evol. 60, 764–773.
Okajima Y., Kumazawa Y. 2010. Mitochondrial genomes of acrodont lizards: Timing of gene rearrangements and phylogenetic and biogeographic implications. BMC Evol. Biol. 10, 141–156.
Amer S.A.M., Kumazawa Y. 2005. Mitochondrial genome of Pogona vitticeps (Reptilia: Agamidae): Control region duplication and the origin of Australian agamids. Gene. 346, 249–256.
Amer S.A.M., Kumazawa Y. 2008. Timing of a mtDNA gene rearrangement and interconcontinental dispersal of varanid lizards. Gene Genet. Syst. 83, 275–280.
Sammler S., Bleidorn C., Tiedemann R. 2011. Full mitochondrial genome sequences of two endemic Philippine hornbill species (Aves: Bucerotidae) provide evidence for pervasive mtDNA recombination. BMC Genomics. 12, 35.
Chan K.M.A., Levin S.A. 2005. Leaky prezygotic isolation and porous genomes: Rapid introgression of maternally inherited DNA. Evolution. 59, 720–729.
Bensasson D., Zhang D.-X., Hartl D.L., Hewitt G.M. 2001. Mitochondrial pseudogenes: Evolution’s misplaced witnesses. Trends Ecol. Evol. 16, 314–322.
Hazkani-Covo E., Zeller R.M., Martin W. 2010. Molecular poltersgeites: Mitochondrial DNA copies (numts) in sequenced nuclear genome. PLOS Genet. 6, e10000834.
Gaziev A.I., Shaikaev G.O. 2010. Nuclear mitochondrial pseudogenes. Mol. Biol. (Moscow). 44, 358–368.
Sunnick P., Hales D.F. 1996. Numerous transposed sequences of mitochondrial cytochrom oxidase I–II in aphids of the genus Sitobion (Hemiptera: Aphydae). Mol. Biol. Evol. 13, 510–524.
Woischnic M., Moraes C.T. 2002. Pattern of organization of human mitochondrial pseudogenes in the nuclear genome. Genome Res. 12, 885–893.
Lopez P., Yuhki N., Masuda R., Modi W., O’Brien S.J. 1994. Numt, a recent transfer and tandem amplification of mitochondrial DNA to the nuclear genome of the domestic cat. J. Mol. Evol. 39, 174–190.
Zhang D.-X., Hewitt G.M. 1996. Nuclear integrations: Challenges for mitochondrial DNA markers. Trends Ecol Evol. 11, 247–251.
Greenwood A., Paabo S. 1999. Nuclear insertion sequences of mtDNA predominant in haire but not in blood of elephants. Mol. Ecol. 8, 133–137.
Mishmar D., Ruiz-Pesini E., Brandon M., Wallace D.C. 2004. Mitochondrial DNA-like sequences in the nucleus (Numt): Insights into our African origins and the mechanism of foregn DNA integration. Hum. Mutat. 23, 125–133.
Triant D.A., DeWoody J.A. 2007. The occurrence, detection, and avoidance of mitochondrial DNA translocations in mammalian systematics amd phylogeography. J. Mammal. 88, 908–920.
Dorner M., Altmann M., Paabo S., Morl M. 2001. Evidence for import of lysyl-tRNA into marsupial mitochondria. Mol. Biol. Cell. 12, 2688–2698.
Funes S., Davidson E., Claros M.G., van List R., Perez-Martinez X., Varquez-Acevedo M., King M.P., Gonzalez-Halphen D. 2002. The typically mitochondrial DNA-encoded ATP6 subunit of the F1F0ATPase is encoded by a nuclear gene in Chlamidomonas reinhardtii. J. Biol. Chem. 277, 6051–6058.
Miraldo A., Hewitt G.M., Dear P.H., Paulo D.S., Emerson B.C. 2012. Numts help us to reconstruct the demographic history of the ocellated lizard (Lacreta lepida) in a secondary contact zone. Mol. Ecol. 21, 1005–1018.
Podnar M., Haring E., Pinsker W., Mayer W. 2007. Unusual origin of a nuclear pseudogene in the Iberian wall lizard: Intergenomic and interspecific transfer of a large section of the mitochondrial genome in the genus Podarcis (Lacertidae). J. Mol. Evol. 64, 308–320.
Kizirian D., Trager A., Donnelly M.A., Wright J.W. 2004. Evolution of Galapagos Island lava lizards (Iguania: Tropiduridae: Microlophus). Mol. Phyl. Evol. 32, 761–769.
Jesus J., Harris D.J., Brehm A. 2005. Phylogeography of Mabuya maculilabris (Reptilia) from Sao Tome Island (Gulf of Guinea) inferred from mtDNA sequences. Mol. Phyl. Evol. 37, 503–510.
Steinfartz S., Glaberman S., Lanterbecq D., Russello M.A., Rosa S., Hanley T C., Marquez C., Snell H.L., Snell H.M., Gentile G., Dell’Olmo G., Powell A.M., Caccone A. 2009. Progressive colonization and restricted gene flow shape island-dependent population structure in Galapagos marine iguanas (Amblyrhynchus cristatus). BMC Evol. Biol. 9, 297–315.
Poulakakis N., Lymberakis P., Valakos E., Zouros E., Mylonas M. 2005. Phylogenetic relationships and biogeography of Podarcis species from the Balkan Peninsula, by Bayesian and maximum likelihood analyses of mitochondrial DNA sequences. Mol. Phyl. Evol. 37, 845–857.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © V.V. Grechko, 2013, published in Molekulyarnaya Biologiya, 2013, Vol. 47, No. 1, pp. 61-82.
Rights and permissions
About this article
Cite this article
Grechko, V.V. The problems of molecular phylogenetics with the example of squamate reptiles: Mitochondrial DNA markers. Mol Biol 47, 55–74 (2013). https://doi.org/10.1134/S0026893313010056
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893313010056