Abstract
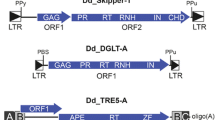
The repeat-induced point mutation mechanism (RIP) is the most intriguing among the known mechanisms of homology-dependent gene inactivation (silencing) because of its ability to produce irreversible mutations in repetitive DNA sequences. Discovered for the first time in Neurospora crassa, RIP is characterized by C:G to T:A transitions in duplicated sequences. The mechanisms and range of occurrence of RIP are still poorly understood. Mobile elements, including retrotransposons, are a common target for the processes that lead to homology-dependent silencing because of their ability to propagate themselves. Comparative analysis of LTR retrotransposons was performed throughout the genomes of two ascomycetes, Aspergillus fumigatus and A. nidulans. “De-RIP” retroelements were reconstructed on the basis of several copies. CpG, CpA, and TpG sites, which are potential targets for mutagenesis, were found at a much lower frequency in mobile elements than in structural genes. The dinucleotide targets of the two species are affected by RIP at different frequencies: mutagenesis occurs at both CpG and CpA sites in A. fumigatus and is confined to CpG dinucleotides in A. nidulans. This work provides a theoretical background for planning the experimental investigation of RIP inactivation in aspergilli.
Similar content being viewed by others
References
Bird A.P. 1986. CpG-rich islands and the function of DNA methylation. Nature. 321, 209–213.
Gowher H., Leismann O., Jeltsch A. 2000. DNA of Drosophila melanogaster contains 5-methylcytosine. EMBO J. 19, 6918–6923.
Vanyushin B.F. 2006. DNA methylation in plants. Curr. Top. Microbiol. Immunol. 301, 67–122.
Doerfler W. 1983. DNA methylation and gene activity. Annu. Rev. Biochem. 52, 93–124.
Norris D.P., Patel D., Kay G.F., Penny G.D., Brockdorff N., Sheardown S.A., Rastan S. 1994. Evidence that random and imprinted Xist expression is controlled by preemptive methylation. Cell. 77, 41–51.
Lloyd V. 2000. Parental imprinting in Drosophila. Genetica. 109, 35–44.
Busslinger M., Hurst J., Flavell R.A. 1983. DNA methylation and the regulation of globin gene expression. Cell. 34, 197–206.
Li E., Bestor T.H., Jaenisch R. 1992. Targed mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 69, 915–926.
Baylin S.B., Herman J.G., Graff J.R., Vertino P.M., Issa J.-P. 1998. Alterations in DNA methylation: A fundamental aspect of neoplasia. Adv. Cancer Res. 72, 141–196.
Bird A.P. 1980. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 8, 1499–1504.
Kricker M.C., Drake J.W., Radman M. 1992. Duplication-targeted DNA methylation and mutagenesis in the evolution of eukaryotic chromosomes. Proc. Natl. Acad. Sci. USA. 89, 1075–1079.
Duncan B.K., Miller J.H. 1980. Mutagenic deamination of cytosine residues in DNA. Nature. 287, 560–561.
Matzke M.A., Matzke A.J. 1998. Epigenetic silencing of plant transgenes as a consequence of diverse cellular defense responses. Cell. Mol. Life Sci. 54, 94–103.
Meyer P., Heidmann I. 1994. Epigenetic variants of a transgenic petunia line show hypermethylation in transgene DNA: An indication for specific recognition of foreign DNA in transgenic plants. Mol. Gen. Genet. 243, 390–399.
Galagan J.E., Selker E.U. 2004. RIP: The evolutionary cost of genome defense. Trends Genet. 20, 417–423.
Faugeron G. 2000. Diversity of homology-dependent gene silencing strategies in fungi. Curr. Opin. Microbiol. 3, 144–148.
Selker E.U. 1999. Epigenetic phenomena in filamentous fungi: Useful paradigms or repeated-induced confusion. Trends Genet. 13, 296–301.
Colot V., Rossingol J.-L. 1999. Eukaryotic DNA methylation as an evolutionary device. BioEssays. 21, 402–411.
Kidwell M.G., Lisch D.R. 2001. Perspective: Transposable elements, parasitic DNA, and genome evolution. Evolution. 55, 1–24.
Kidwell M.G. 2002. Transposable elements and the evolution of genome size in eukaryotes. Genetica. 115, 49–63.
Hua-Van A., Le Rouzic A., Maisonhaute C., Capy P. 2005. Abundance, distribution and dynamics of retrotransposable elements and transposons: similarities and differences. Cytogenet. Genome Res. 110, 426–440.
Novikova O., Fet B., Blinov A. 2007. LOR retrotransposons in the Aspergillus fumigatus and A. nidulans genomes. Mol. Biol. 41, 756–763.
Neuveglise C., Sarfati J., Latge J.-P., Paris S. 1996. Afut1, a retrotransposon-like element from Aspergillus fumigatus. Nucleic Acids Res. 24, 1428–1434.
Paris S., Latge J.P. 2001. Afut2, a new family of degenerate gypsy-like retrotransposon from Aspergillus fumigatus. Med. Mycol. 39, 195–198.
Nielsen M.L., Hermansen T.D., Aleksenko A. 2001. A family of DNA repeats in Aspergillus nidulans has assimilated degenerated retrotransposons. Mol. Genet. Genomics. 265, 883–887.
Thompson J.D., Higgins D.G., Gibson T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 22, 4673–4680.
Kricker M.C., Drake J.W., Radman M. 1992. Duplication-targeted DNA methylation and mutagenesis in the evolution of eukaryotic chromosomes. Proc. Natl. Acad. Sci. USA. 89, 1075–1079.
Cambareri E.B., Aisner R., Carbon J. 1998. Structure of the chromosome VII centromere region in Neurospora crassa: Degenerate transposons and simple repeats. Mol. Cell. Biol. 18, 5465–5477.
Daboussi M.J., Daviere J.M., Graziani S., Langin T. 2002. Evolution of the Fot1 transposons in the genus Fusarium: Discontinuous distribution and epigenetic inactivation. Mol. Biol. Evol. 19, 510–520.
Aleksenko A., Gems D., Clutterbuck J. 1996. Multiple copies of MATE elements support autonomous plasmid replication in Aspergillus nidulans. Mol. Microbiol. 20, 427–434.
Geiser D.M., Timberlake W.E., Arnold M.L. 1996. Loss of meiosis in Aspergillus. Mol. Biol. Evol. 13, 809–817.
Poggeler S. 2002. Genomic evidence for mating abilities in the asexual pathogen Aspergillus fumigatus. Curr Genet. 42, 153–160.
Varga J. 2003. Mating type gene homologues in Aspergillus fumigatus. Microbiology. 149, 816–819.
Paoletti M., Rydholm C., Schwier E.U., Anderson M.J., Szakacs G., Lutzoni F., Debeaupuis J.P., Latge J.P., Denning D.W., Dyer P.S. 2005. Evidence for sexuality in the opportunistic fungal pathogen Aspergillus fumigatus. Curr Biol. 15, 1242–1248.
Gow N.A., Brown A.J., Odds F.C. 2000. Candida’ arranged marriage. Science. 289, 256–257.
Tamame M., Antequera F., Villanueva J.R., Santos T. 1983. High-frequency conversion to a “fluffy” developmental phenotype in Aspergillus spp. by 5-azacytidine treatment: Evidence for involvement of a single nuclear gene. Mol. Cell. Biol. 3, 2287–2297
Gowher H., Ehrlich K.C., Jeltsch A. 2001. DNA from Aspergillus flavus contains 5-methylcytosine. FEMS Microbiol. Lett. 205, 151–155.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © O.S. Novikova, V. Fet, A.G. Blinov, 2007, published in Molekulyarnaya Biologiya, 2007, Vol. 41, No. 6, pp. 973–981.
Rights and permissions
About this article
Cite this article
Novikova, O.S., Fet, V. & Blinov, A.G. Homology-dependent inactivation of LTR retrotransposons in Aspergillus fumigatus and A. nidulans genomes. Mol Biol 41, 886–893 (2007). https://doi.org/10.1134/S0026893307060039
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1134/S0026893307060039