Abstract
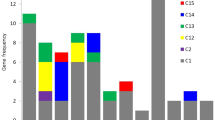
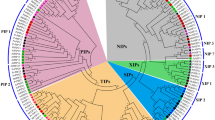
Peptidases occur naturally in all organisms and their genes comprise 1–5% of the total number of genes. Genetic, biochemical, and molecular approaches used in recent years led to the identification and characterization of several plant organelle proteases, all of them being homologous to bacterial proteases best characterized in Escherichia coli. Here we report isolating and characterizing three novel genes, namely Sszn-mp1, Sszn-mp2, and SsZn-mp3 from Solanum surattense. To identify the subcellular location, structures, and functions of these three genes, integrated genomic approaches of data mining, expression profiling, and bioinformatic predictions were used. Sszn-mp is found to be constitutively expressed in tissues and regulated by various stimuli. Analysis of eight zinc-metalloproteases (Zn-MPs) deduced or assembled from Arabidopsis thaliana, tomato, potato, cotton, barley, sugarcane, and rice and four Zn-MPs from cyanobacteria (blue-green algae) in the GenBank database reveals that these proteins belong to a novel conserved membrane zinc-metalloprotease family. The plant Zn-MP members share more than 62% overall identity with SsZn-MP3, whereas four putative ATP-dependent zinc-proteases of cyanobacteria have low identity with SsZn-MP3 and their N-termini are about 110 amino acids shorter than those of plant Zn-MPs. The Zn-MP homologous sequences are found neither in other eukaryotic nor prokaryotic databases, suggesting that this family is specific to plants and cyanobacteria. The plant Zn-MP genes encoding membrane proteins are potentially targeted to chloroplast and plasma membranes, and the bacterial Zn-MPs are targeted to the cytoplasmic membrane, and their N-terminal targeting peptides are cleaved off for targeting the mature proteins to their subcellular compartments. The Zn-MP proteins contain a conserved zinc-binding site (HEAGHX19E/DX46∼48EX7E), a potential G-protein coupled receptors family 1 signature, and a triplet motif (N-R/K-F) in plant Zn-MPs, a D/E-R-Y motif in the four bacterial Zn-MPs, suggesting that the different mature forms of Zn-MPs may function as proteases and/or signal receptors.
Similar content being viewed by others
Abbreviations
- aa:
-
amino acid
- ER:
-
endoplasmic reticulum
- EST:
-
expressed sequence tag
- GPCR:
-
G-protein coupled receptor
- GPI:
-
glycosylated phosphatidylinositol
- MeJa:
-
methyl jasmonate
- ORF:
-
open reading frame
- PCR:
-
polymerase chain reaction
- RT-PCR:
-
real time PCR
- SA:
-
salicylic acid
- TMD:
-
transmembrane domain
- UTR:
-
untranslated region
- Zn-MP:
-
zinc-metalloprotease
- GrZn-MP:
-
Zn-MP from Gossipium raimondii
References
Moberg, P., Stahl, A., Bhushan, S., Wright, S.J., Eriksson, A., Bruce, B.D., and Glaser, E., Characterization of a Novel Zinc Metalloprotease Involved in Degrading Targeting Peptides in Mitochondria and Chloroplasts, Plant J., 2003, vol. 36, pp. 616–628.
Hedstrom, L., Serine Protease Mechanism and Specificity, Chem. Rev., 2002, vol. 102, pp. 4501–4523.
Southan, C., A Genomic Perspective on Human Proteases as Drug Targets, Drug Discov. Today, 2001, vol. 6, pp. 681–688.
Puente, X.S. and Lopez-Otin, C., A Genomic Analysis of Rat Proteases and Protease Inhibitors, Genome Res., 2004, vol. 14, pp. 609–622.
Rawlings, N.D., Tolle, D.P., and Barrett, A.J., MEROPS: The Peptidase Database, Nucleic Acids Res., 2004, vol. 32, pp. D160–D164.
Schaller, A., A Cut above the Rest: The Regulatory Function of Plant Proteases, Planta, 2004, vol. 220, pp. 183–197.
Gomis-Ruth, F.X., Structural Aspects of the Metzincin Clan of Metalloendopeptidases, Mol. Biotechnol., 2003, vol. 24, pp. 157–202.
Zhang, H.Z., Hackbarth, C.J., Chansky, K.M., and Chambers, H.F., A Proteolytic Transmembrane Signaling Pathway and Resistance to beta-Lactams in Staphylococci, Science, 2001, vol. 291, pp. 1962–1965.
Kheradmand, F. and Werb, Z., Shedding Light on Sheddases: Role in Growth and Development, BioEssay, 2002, vol. 24, pp. 8–12.
Rawlings, N.D. and Barrett, A.J., Evolutionary Families of Metallopeptidases, Proteolytic Enzymes: Aspartic and Metallo Peptidases, Abelson, J.N., Simon, M.I., and Barrett, A.J., Eds., San Diego: Academic, 1995, pp. 183–228.
Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J., Basic Local Alignment Search Tool, J. Mol. Biol., 1990, vol. 215, pp. 403–410.
Benson, D.A., Karsch-Mizrachi, I., Lipman, D.J., Ostell, J., and Wheeler, D.L., GenBank: Update, Nucleic Acids Res., 2004, vol. 32, pp. D23–D26.
Pearson, W.R., Rapid and Sensitive Sequence Comparison with Fastp and Fasta, Nucleic Acids Res., 1990, vol. 183, pp. 63–98.
Thompson, J.D., Higgins, D.G., and Gibson, T.J., Clustal-W—Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice, Nucleic Acids Res., 1994, vol. 22, pp. 4673–4680.
Nakai, K. and Kanehisa, M., A Knowledge Base for Predicting Protein Localization Sites in Eukaryotic Cells, Genomics, 1992, vol. 14, pp. 897–911.
Bannai, H., Tamada, Y., Maruyama, O., Nakai, K., and Miyano, S., Extensive Feature Detection of N-Terminal Protein Sorting Signals, Nucleic Acids Res., 2002, vol. 18, pp. 298–305.
Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G., Predicting Subcellular Localization of Proteins Based on Their N-Terminal Amino Acid Sequence, Nucleic Acids Res., 2000, vol. 300, pp. 1005–1016.
Bendtsen, J.D., Nielsen, H., von Heijne, G., and Brunak, S., Improved Prediction of Signal Peptides: SignalP 3.0, Nucleic Acids Res., 2004, vol. 340, pp. 783–795.
Emanuelsson, O., Nielsen, H., and von Heijne, G., ChloroP, a Neural Network-Based Method for Predicting Chloroplast Transit Peptides and Their Cleavage Sites, Protein Sci., 1999, vol. 8, pp. 978–984.
Claros, M.G. and Vincens, P., Computational Method to Predict Mitochondrially Imported Proteins and Their Targeting Sequences, Eur. J. Biochem., 1996, vol. 241, pp. 779–786.
Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E.L.L., Predicting Transmembrane Protein Topology with a Hidden Markov Model: Application to Complete Genomes, Nucleic Acids Res., 2001, vol. 305, pp. 567–580.
Combet, C., Blanchet, C., Geourjon, C., and Deleage, G., NPS: Network Protein Sequence Analysis, Trends Biol. Sci., 2000, vol. 25, pp. 147–150.
Blom, N., Gammeltoft, S., and Brunak, S., Sequence and Structure-Based Prediction of Eukaryotic Protein Phosphorylation Sites, J. Mol. Biol., 1999, vol. 294, pp. 1351–1362.
Letunic, I., Copley, R.R., Schmidt, S., Ciccarelli, F.D., Doerks, T., Schultz, J., Ponting, C.P., and Bork, P., SMART 4.0: Towards Genomic Data Integration, Nucleic Acids Res., 2004, vol. 32, pp. D142–D144.
Schultz, J., Milpetz, F., Bork, P., and Ponting, C.P., SMART, a Simple Modular Architecture Research Tool: Identification of Signaling Domains, Proc. Natl. Acad. Sci. USA, 1998, vol. 95, pp. 5857–5864.
Marchler-Bauer, A. and Bryant, S.H., CD-Search: Protein Domain Annotations on the Fly, Nucleic Acids Res., 2004, vol. 32, pp. W327–W331.
Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J.H., Zhang, Z., Miller, W., and Lipman, D.J., Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs, Nucleic Acids Res., 1997, vol. 25, pp. 3389–3402.
Durner, J., Shah, J., and Kessig, D.F., Salicylic Acid and Disease Resistance in Plants, Trends Plant Sci., 1997, vol. 2, pp. 266–274.
Dong, X., SA, JA, Ethylene, and Disease in Plants, Curr. Opin. Plant Biol., 1998, vol. 1, pp. 316–323.
Kikuchi, S., Satoh, K., Nagata, T., Kawagashira, N., Doi, K., Kishimoto, N., Yazaki, J., Ishikawa, M., Yamada, H., Ooka, H., Hotta, I., Kojima, K., Namiki, T., Ohneda, E., Yahagi, W., Suzuki, K., Li, C.J., Ohtsuki, K., Shishiki, T., Otomo, Y., Murakami, K., Iida, Y., Sugano, S., Fujimura, T., Suzuki, Y., Tsunoda, Y., Kurosaki, T., Kodama, T., Masuda, H., Kobayashi, M., Xie, Q.H., Lu, M., Narikawa, R., Sugiyama, A., Mizuno, K., Yokomizo, S., Niikura, J., Ikeda, R., Ishibiki, J., Kawamata, M., Yoshimura, A., Miura, J., Kusumegi, T., Oka, M., Ryu, R., Ueda, M., Matsubara, K., Kawai, J., Carninci, P., Adachi, J., Aizawa, K., Arakawa, T., Fukuda, S., Hara, A., Hashizume, W., Hayatsu, N., Imotani, K., Ishii, Y., Itoh, M., Kagawa, I., Kondo, S., Konno, H., Miyazaki, A., Osato, N., Ota, Y., Saito, R., Sasaki, D., Sato, K., Shibata, K., Shinagawa, A., Shiraki, T., Yoshino, M., Hayashizaki, Y., and Yasunishi, A., Collection, Mapping, and Annotation of over 28,000 cDNA Clones from Japonica Rice, Science, 2003, vol. 301, pp. 376–379.
Agarraberes, F.A. and Dice, J.F., Protein Translocation across Membranes, Biochim. Biophys. Acta-Biomembranes, 2001, vol. 1513, pp. 1–24.
Dong, T. and Cutting, S., Sporulation Factor SpoIVFB, Handbook of Proteolytic Enzymes, Barrett, A., Rawlings, N., and Woessner, J., Eds., London: Elsevier, 2004, pp. 989–991.
Yu, Y.T.N. and Kroos, L., Evidence that SpoIVFB Is a Novel Type of Membrane Metalloprotease Governing Intercompartmental Communication during Bacillus subtilis Sporulation, J. Bacteriol., 2000, vol. 182, pp. 3305–3309.
Becker, A.B. and Roth, R.A., Identification of Glutamate-169 as the 3rd Zinc-Binding Residue in Proteinase-111, a Member of the Family of Insulin-Degrading Enzymes, Biochem. J., 1993, vol. 292, pp. 137–142.
Perlman, R.K., Gehm, B.D., Kuo, W.L., and Rosner, M.R., Functional Analysis of Conserved Residues in the Active Site of Insulin-Degrading Enzyme, J. Biol. Chem., 1993, vol. 268, pp. 21538–21544.
Perlman, R.K. and Rosner, M.R., Identification of Zinc Ligands of the Insulin-Degrading Enzyme, J. Biol. Chem., 1994, vol. 269, pp. 33140–33145.
Cole, S.T., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D., Gordon, S.V., Eiglmeier, K., Gas, S., Barry, C.E. III, Tekaia, F., Badcock, K., Basham, D., Brown, D., Chillingworth, T., Connor, R., Davies, R., Devlin, K., Feltwell, T., Gentles, S., Hamlin, N., Holroyd, S., Hornsby, T., Jagels, K., Krogh, A., McLean, J., Moule, S., Murphy, L., Oliver, K., Osborne, J., Quail, M.A., Rajandream, M.-A., Rogers, J., Rutter, S., Seeger, K., Skelton, J., Squares, R., Squares, S., Sulston, J.E., Taylor, K., Whitehead, S., and Barrell, B.G., Deciphering the Biology of Mycobacterium tuberculosis from the Complete Genome Sequence, Nature, 1998, vol. 393, pp. 537–544.
Strosberg, A.D., Structure-Function Relationship of Proteins Belonging to the Family of Receptors Coupled to GTP-Binding Proteins, Eur. J. Biochem., 1991, vol. 196, pp. 1–10.
Horn, F., Bettler, E., Oliveira, L., Campagne, F., Cohen, F.E., and Vriend, G., GPCRDB Information System for G Protein-Coupled Receptors, Nucleic Acids Res., 2003, vol. 31, pp. 294–297.
Chung, D.A., Wade, S.M., Fowler, C.B., Woods, D.D., Abada, P.B., Mosberg, H.L., and Neubig, R.R., Mutagenesis and Peptide Analysis of the DRY Motif in the Alpha 2A Adrenergic Receptor: Evidence for Alternate Mechanisms in G Protein-Coupled Receptors, Biochem. Biophys. Res. Commun., 2002, vol. 293, pp. 1233–1241.
Alewijnse, A.E., Timmerman, H., Jacobs, E.H., Smit, M.J., Roovers, E., Cotecchia, S., and Leurs, R., The Effect of Mutations in the DRY Motif on the Constitutive Activity and Structural Instability of the Histamine H-2 Receptor, Mol. Pharmacol., 2000, vol. 57, pp. 890–898.
Mhaouty-Kodja, S., Barak, L.S., Scheer, A., Abuin, L., Diviani, D., Caron, M.G., and Cotecchia, S., Constitutively Active alpha-1b Adrenergic Receptor Mutants Display Different Phosphorylation and Internalization Features, Mol. Pharmacol., 1999, vol. 55, pp. 339–347.
Devoto, A., Piffanelli, P., Nilsson, I., Wallin, E., Panstruga, R., von Heijne, G., and Schulze-Lefert, P., Topology, Subcellular Localization, and Sequence Diversity of the Mlo Family in Plants, J. Biol. Chem., 1999, vol. 274, pp. 34993–35004.
Plakidou-Dymock, S., Dymock, D., and Hooley, R., A Higher Plant Seven-Transmembrane Receptor That Influences Sensitivity to Cytokinins, Curr. Biol., 1998, vol. 8, pp. 315–324.
Josefsson, L.G. and Rask, L., Cloning of a Putative G-Protein-Coupled Receptor from Arabidopsis thaliana, Eur. J. Biochem., 1997, vol. 249, pp. 415–420.
Colucci, G., Apone, F., Alyeshmerni, N., Chalmers, D., and Chrispeels, M.J., GCR1, the Putative Arabidopsis G Protein-Coupled Receptor Gene Is Cell Cycle-Regulated, and Its Overexpression Abolishes Seed Dormancy and Shortens Time to Flowering, Proc. Natl. Acad. Sci. USA, 2002, vol. 99, pp. 4736–4741.
Author information
Authors and Affiliations
Additional information
Published in Russian in Fiziologiya Rastenii, 2007, Vol. 54, No. 1, pp. 73–84.
The text was submitted by the authors in English.
Rights and permissions
About this article
Cite this article
Sun, X., Liu, X., Guo, S. et al. Cloning and analysis of a novel conserved membrane zinc-metalloprotease family from Solanum surattense . Russ J Plant Physiol 54, 63–73 (2007). https://doi.org/10.1134/S1021443707010104
Received:
Issue Date:
DOI: https://doi.org/10.1134/S1021443707010104