Abstract
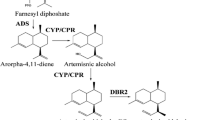
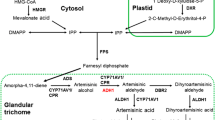
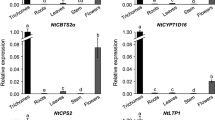
Artemisinin, a sesquiterpene lactone endoperoxide derived from Artemisia annua L.(Asteraceae), is the most effective antimalarial drug. We used two methods: genome walking and thermal asymmetric interlaced polymerase chain reaction, to isolate the unknown 5′-flanking sequence of the cyp71av1 gene. The subsequent sequence analysis using bio informatics software revealed that there are several cis-acting elements inside the cyp71av1 promoter. The 5′-rapid amplification of the cDNA ends method was used to determine the transcription start site of the cyp71av1 gene. We then mapped it at the 18 base upstream of the ATG initiation codon. For simple functional characterization, we built fusion vectors between the 5′-deletion promoter and the gus reporter gene. The expression levels of the transferred vectors into A. annua L. were analyzed by the transient expression way. The β-glucuronidase assay results indicated that deletion of the region to −1551 bp did not lead to much damage in the GUS activity, whereas further deletion, to −1155 bp, resulted in a 5.5-fold reduction of GUS activity. In stabilized transgenic A. annua L. seedlings we observed that GUS expression was restricted to trichomes, which means that the promoter of the cyp71av1 gene is trichome-specific. Compared with the constitutive CaMV 35S promoter, which can express genes throughout the plant, influence on the trichome system through the trichome-specific expression promoter merely imperils plant growth. In addition, the promoter of the cyp71av1 gene contains several binding sites for transcription factors, which implies that the cyp71av1 promoter responds to more than one form of stimulation.
Similar content being viewed by others
Abbreviations
- 6-BA:
-
6-benzylaminopurine
- NAA:
-
naphthaleneacetic acid
- SA:
-
salicylic acid
- MU:
-
methyl umbelliferone
- AS:
-
acetasyringone
References
WHO. 2005. World Malaria Report.
Tianwei Liu, Lingbo Qu, Bingren Xiang. 2003. The progress of artemisinin anti-malaria drugs. Chin. J. Med. Guide. 5, 399–401.
WHO. 2006. Guidelines for the Treatment of Malaria. Geneva: WHO.
Liu C.Z., Zhao Y., Wang Y.C. 2006. Artemisinin: Current state and perspectives for biotechnological production of an antimalarial drug. Appl. Microbiol. Biotechnol. 71, 11–20.
Bertea C.M., Freije J.R., van der Woude H., et al. 2005. Identification of intermediates and enzymes involved in the early steps of artemisinin biosynthesis in Artemisia annua. Planta Med. 71, 40–47.
Teoh K.H., Polichuk D.R., Reed D.W., et al. 2006. Artemisia annua L. (Asteraceae) trichome-specific cDNAs reveal CYP71AV1, a cytochrome P450 with a key role in the biosynthesis of the antimalarial sesquiterpene lactone artemisinin. FEBS Lett. 580, 1411–1416.
Ro D.K., Paradise E.M., Ouellet M., et al. 2006. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 440, 940–943.
Jing F.Y., Zhang L., Li M.Y., et al. 2009. Abscisic acid (ABA) treatment increases artemisinin content in Artemisia annua by enhancing the expression of genes in artemisinin biosynthetic pathway. Biologia. 64, 319–323.
Vergauwe A., Cammaert R., Vandenberghe D., et al. 1996. Agrobacterium tumefaciens-mediated transformation of Artemisia annua L. and regeneration of transgenic plants. Plant Cell Rep. 15, 929–933.
Jefferson R.A., Kavanagh T.A., Bevan M.W. 1987. GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907.
Cote C., Rutledge R.G. 2003. An improved MUG fluorescent assay for the determination of GUS activity within transgenic tissue of woody plants. Plant Cell Rep. 21, 619–624.
Lescot M., Déhais P., Moreau Y., et al. 2002. Plant-CARE: A database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327.
Higo K., Ugawa Y., Iwamoto M., Korenaga T. 1999. SIGNAL SCAN: A computer program that scans DNA sequences for eukaryotic transcriptional elements. CABIOS. 7, 203–206.
Ke J., Choi J.K., Smith M., et al. 1997. Structure of the CAC1 gene and in situ characterization of its expression (the Arabidopsis thaliana gene coding for the biotin-containing subunit of the plastidic acetyl-coenzyme a carboxylase). Plant Physiol. 113, 357–365.
Menkens A.E., Schindler U., Cashmore A.R. 1995. The G-box: A ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem. Sci. 20, 506–510.
Guiltinan M.J., Marcotte W.R.J., Quatrano R.S. 1990. A plant leucine zipper protein that recognizes an abscisic acid response element. Science. 250, 267–271.
McKendree W.L.J., Ferl R.J. 1992. Functional elements of the Arabidopsis Adh promoter include the G-box. Plant Mol. Biol. 19, 859–862.
Block A., Dangl J.L., Hahlbrock K., et al. 1990. Functional borders, genetic fine structure, and distance requirements of cis elements mediating light responsiveness of the parsley chalcone synthase promoter. Proc. Natl. Acad. Sci. U. S. A. 87, 5387–5391.
Fujita Y., Fujita M., Satoh R., et al. 2005. AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell. 17, 3470–3488.
Hobo T., Asada M., Kowyama Y., Hattori T. 1999. ACGT-containing abscisic acid response element (ABRE) and coupling element 3 (CE3) are functionally equivalent. Plant J. 19, 679–689.
Mason H.S., DeWald D.B., Mullet J.E. 1993. Identification of a methyl jasmonate-responsive domain in the soybean vspB promoter. Plant Cell. 5, 241–251.
Baker S.S., Wilhelm K.S., Thomashow M.F. 1994. The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 24, 701–713.
Urao T., Yamaguchi-Shinozaki K., Urao S., Shinozaki K. 1993. An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell. 5, 1529–1539.
Author information
Authors and Affiliations
Corresponding author
Additional information
The article is published in the original.
Rights and permissions
About this article
Cite this article
Wang, Y., Yang, K., Jing, F. et al. Cloning and characterization of trichome-specific promoter of cpr71av1 gene involved in artemisinin biosynthesis in Artemisia annua L.. Mol Biol 45, 751–758 (2011). https://doi.org/10.1134/S0026893311040145
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893311040145