Abstract
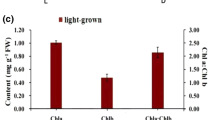
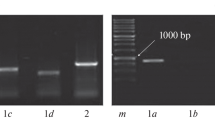
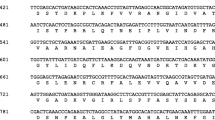
We report the characterisation of two cytochrome b5 genes and their spatial and temporal patterns of expression during development in olive, Olea europaea. A PCR-generated probe, based on a tobacco cytochrome b5 sequence, was used to isolate two full-length cDNA clones (cytochrome b5-15 and cytochrome b5-38) from a library derived from 13 WAF olive fruits. The cDNAs encoded proteins of 17.0 and 17.7 kDa, which contained all the characteristic motifs of cytochromes b5 from other organisms and exhibited 63% identity and 85% similarity with each other. The olive cytochrome b5-15 cDNA was then used as a probe for more detailed analysis. Southern blotting revealed a gene family of at least 4–6 members while northern blotting and in situ hybridisation showed a highly specific pattern of gene expression. Very low levels of cytochrome b5 mRNA were detected in tissues characterised by high rates of lipid accumulation, such as young expanding leaves, maturing seeds and ripening mesocarp. The cytochrome b5 genes were not induced at 6 °C and their response to ABA was relatively slow compared with fatty acid desaturase genes. In contrast, high levels of cytochrome b5 gene expression were found in young fruits at the pattern formation (globular/heart) stage of embryogenesis and in vascular and transmitting tissues of male and female reproductive organs. The data are consistent with a major role for cytochrome b5 in developmental processes related to plant reproduction in addition to being an electron donor to microsomal desaturases.
Similar content being viewed by others
References
Bafor NL, Smith MA, Jonsson L, Stobart K, Stymne S: Biosynthesis of vernoleate (cis-12-epoxyoctadeca-cis-9-enoate) in microsomal preparations from developing endosperm of Euphorbia lagascae. Arch Biochem Biophys 303: 145–151 (1993).
Berberich T, Harada M, Sugawara K, Kodama H, Iba K, Karuso T: Two maize genes encoding omega-3 fatty acid desaturase and their differential expression to temperature. Plant Mol Biol 36: 297–306 (1998).
Bolwell GP, Bozak K, Zimmerlin A: Plant cytochrome P450. Phytochemistry 37: 1491–1506 (1994).
Bonnerot C, Galle AM, Jolliot A, Kader J-C: Purification and properties of plant cytochrome b5. Biochem J 226: 331–334 (1985).
Choe S, Dilkes BP, Fujioka S, Takatsuto S, Sakurai A, Feldmann KA: The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22α hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 10: 231–243 (1998).
Church G, Gilbert W: Genomic sequencing. Proc Natl Acad Sci USA 81: 1991–1995 (1985).
Demirkapi N, Carreau JP, Ghesquier D: Evidence against cytochrome b5 involvement in liver microsomal fatty acid elongation. Biochim Biophys Acta 1082: 49–56 (1991).
Donaldson RP, Luster DG: Multiple forms of plant cytochromes P450. Plant Physiol 96: 669–674 (1991).
Haralampidis K, Milioni D, Sanchez J, Baltrusch M, Heinz E, Hatzopoulos P: Temporal and transient expression of stearoyl-ACP carrier protein desaturase gene during olive fruit development. J Exp Bot 49: 1661–1669 (1998).
Heppard EP, Kinney AJ, Stecca KI, Miao G-H: Developmental and growth temperature regulation of two different microsomal omega-6 desaturase genes in soybeans. Plant Physiol 110: 311–3l9 (1996).
Honjo K, Ishibashi T, Imai Y: Partial purification and characterization of lathosterol 5-desaturase from rat liver microsomes. J Biochem 97: 955–959 (1985).
Imai Y: The roles of cytochrome b5 in reconstituted monooxygenase systems containing various forms of hepatic microsomal cytochrome P-450. J Biochem 89: 351–362 (1981).
Jackson DP: In situ hybridization in plants. In: Bowles DJ, Gurr SJ, McPheron M. <nt>(Eds)</nt> Molecular Plant Pathology: A Practical Approach, Oxford University Press, Oxford, pp. 140–154 (1991).
Jollie DR, Sligar SG, Schuler M: Purification and characterization of microsomal cytochrome b5 and NADH cytochrome b5 reductase from Pisum sativum. Plant Physiol 85: 457–462 (1987).
Katagiri M, Kagawa N, Waterman MR: The role of cytochrome b5 in the biosynthesis of androgens by human P450c17. Arch Biochem Biophys 317: 343–347 (1995).
Kearns EV, Keck P, Somerville CR: Primary structure of cytochrome b5 from cauliflower (Brassica oleracea L.) deduced from peptide and cDNA sequences. Plant Physiol 99: 1254–1257 (1992).
Kearns EV, Hughly S, Somerville CR: The role of cytochrome b5 in delta 12 desaturation of oleic acid by microsomes of safflower (Carthamus tinctorius L.). Arch Biochem Biophys 284: 431–436 (1991).
Kuroda R, Kinoshita J, Honsho M, Mitoma J, Ito A: In situ topology of cytochrome b5 endoplasmic reticulum membrane. J Biochem 120: 828–833 (1996).
Lederer F: The cytochrome b5-fold: an adaptable module. Biochimie 76: 674–692 (1994).
Mathews FS: The structure, function and evolution of cytochromes. Prog Biophys Mol Biol 45: 1–56 (1985).
McCormick S: Male gametophyte development. Plant Cell 5: 1265–1275 (1993).
Mitchell AG, Martin CE: A novel cytochrome b5-like domain is linked to the carboxyl terminus of the Saccharomyces cerevisiae delta-9 fatty acid. J Biol Chem 270: 29766–29772 (1995).
Mizutani M, Ohta D: Two isoforms of NADPH:cytochrome P450 reductase in Arabidopsis thaliana. Plant Physiol 116: 357–367 (1998).
Murashige T, Skoog F: A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol Plant 15: 473–497 (1962).
Murphy DL, Woodrow IE, Mukherjee KD, Latzko E: Solubilisation of oleoyl-CoA thioesterase, oleoyl-CoA phosphatidylcholine acyltransferase and oleolylphosphatidylycholine desaturase. FEBS Lett 162: 442–446 (1983).
Murphy DJ, Piffanelli P: Fatty acid desaturases: structure, mechanism and regulation. In: Harwood JL <nt>(ed)</nt> Plant Lipid Biosynthesis: Recent Advances of Agricultural Importance, Cambridge University Press, Cambridge, UK, in press (1998).
Murray MG, Thompson WF: Rapid isolation of high weight plant DNA. Nucl Acids Res 8: 4321–4325 (1980).
Napier JA, Sayanova O, Stobart AK, Shewry PR: A new class of cytochrome b5 fusion proteins. Biochem J 328: 717–718 (1997).
Napier JA, Hey SJ, Lacey DJ, Shewry PR: Identification of a Caenorhabditis elegans delta-6-fatty-acid-desaturase by heterologous expression in Saccharomyces cerevisiae. Biochem J 330: 611–614 (1998).
Napier JA, Smith MA, Stobart AK, Shrewy PR: Isolation of a cDNA encoding a cytochrome b5 specifically expressed in developing tobacco seeds. Planta 197: 200–202 (1995).
Ohba M, Sato R, Yoshida Y, Bieglmayer C, Ruis H: Mutant and immunochemical studies on the involvement of cytochrome b5 in fatty acid desaturation by yeast microsomes. Biochim Biophys Acta 572: 352–362 (1979).
Ozols J: Structure of cytochrome b5 and its topology in the microsomal membrane. Biochim Biophys Acta 997: 121–130 (1989).
Paltuaf F, Prough RA, Masters BS, Johnston JM: Evidence for the partipication of cytochrome b5 in plasmalogen biosynthesis. J Biol Chem 249: 2661–2662 (1974).
Piffanelli P, Ross JHE, Murphy DJ: Biogenesis and function of the lipidic structures of pollen grains. Sex Plant Reprod 11: 65–80 (1998).
Piffanelli P, Ross JHE, Murphy DJ: Intra and extracellular lipid composition and associated gene expression patterns during pollen development in Brassica napus. Plant J 11: 549–562 (1997).
Poghosyan ZP, Haralampidis K, Martsinkovskaya AI, Murphy DJ, Hatzopoulos P: Developmental regulation and spatial expression of an ώ-3 fatty acid desaturase from Olea europaea. Plant Physiol Biochem, in press (1999).
Rodriguez-Maranon MJ, Qui F, Stark RE, White SP, Zhang X, Foundling SI, Rodriguez V, Schilling CI, Bunce RAI, Rivera M: 13CNMRspectroscopic and X-ray crystallographic study of the role played by mitochondrial cytochrome b5 heme propionates in the electrostatic binding to cytochrome c. Biochemistry 35: 16378–16390 (1996).
Roy D, Strobel HW, Liehr JG: Cytochrome b5-mediated redox cycling of estrogen. Arch Biochem Biophys 285: 331–338 (1991).
Sambrook J, Fritsch EF, Maniatis T: Molecular Cloning: A Laboratory Manual, 2nd ed., Cold Spring Harbor Laboratory Press. Cold Spring Harbor, NY (1989).
Sanger F, Nicklen S, Coulson AR: DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74: 5463–5467 (1977).
Sayanova O, Smith MA, Lapinskas P, Stobart AK, Dobson G, Christie WW, Shewry PR, Napier J: Expression of a borage desaturase cDNA containing an N-terminal cytochrome b5 domain results in the accumulation of high levels of delta-6-desaturated fatty acids in transgenic tobacco. Proc Natl Acad Sci USA 94: 4211–4216 (1997).
Scott R, Dagless E, Hodge R, Wyat P, Soufleri I, Draper J: Patterns of gene expression in developing anthers of Brassica napus. Plant Mol Biol 17: 195–207 (1991).
Smith MA, Stobart, AK, Shewry JR, Napier JA: Tobacco cytochrome b5: cDNA isolation, expression analysis and in vitro protein targeting. Plant Mol Biol 25: 527–537 (1994).
Smith MA, Cross AR, Jones QT, Griffiths WT, Stymne, S, Stobart K: Electron-transport components of the 1-acyl-2-oleoylsn-glycero-3-phosphocholine delta 12-desaturase (delta 12-desaturase) in microsomal preparations from developing saf-flower (Carthamus tinctorius L.) cotyledons. Biochem J 272: 23–29 (1990).
Smith, MA, Jonsson L, Stymne S, Stobart K: Evidence for cytochrome b5 as an electron donor in ricinoleic acid biosynthesis in microsomal preparations from developing castor bean (Ricinus communis L.). Biochem J 287: 141–144 (1992).
Sperling P, Schmidt H, Heinz E: A cytochrome-b5-containing fusion protein similar to plant acyl lipid desaturases. Eur J Biochem 232: 798–805 (1995).
Takeshita M, Tamura M, Yoshida S, Yubisui T: Palmitoyl-CoA elongation in brain microsomes: dependence on cytochrome b5 and NADH-cytochrome b5 reductase. J Neurochem 45: 1390–1395 (1985).
Vergeres G, Waskell L: Cytochrome b5, its functions, structure and membrane topology. Biochimie 77: 604–620 (1995).
Yamazaki K, Johnson WW, Ueng Y-F, Shimada T, Guengerich FP: Lack of electron transfer from cytochrome b5 in stimulation of catalytic activities of cytochrome P450 3A4. J Biol Chem 271: 27438–27444 (1996).
Yamazaki H, Ueng Y-F, Shimada T, Guengerich FP: Roles of divalent metal ions in oxidations catalyzed by recombinant cytochrome P450 3A4 and replacement of NADPH-cytochrome P450 reductase with other flavoproteins, ferredoxin, and oxygen surrogates. Biochemistry 34: 8380–8389 (1995).
Yang MX, Cederbaum AI: Role of cytochrome b5 in NADH-dependent microsomal reduction of ferric complexes, lipid peroxidation, and hydrogen peroxide generation. Arch Biochem Biophys 324: 282–292 (1995).
Yokota T: The structure, biosynthesis and function of brassinosteroids. Trends Plant Sci 2: 137–143 (1997).
Zhang M, Scott JG: Cytochrome b5 involvement in cytochrome P450 monooxygenase activities in house fly microsomes. Arch Insect Biochem Physiol 27: 205–216 (1994).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Martsinkovskaya, A.I., Poghosyan, Z.P., Haralampidis, K. et al. Temporal and spatial gene expression of cytochrome B5 during flower and fruit development in olives. Plant Mol Biol 40, 79–90 (1999). https://doi.org/10.1023/A:1026417710320
Issue Date:
DOI: https://doi.org/10.1023/A:1026417710320