Abstract
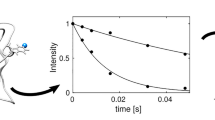
Comparatively small molecules such as peptides can show a high internal mobility with transitions between several conformational minima and sometimes coupling between rotational and internal degrees of freedom. In those cases the interpretation of NMR relaxation data is difficult and the use of standard methods for structure determination is questionable. On the other hand, in the case of those system sizes, the timescale of both rotational and internal motions is accessible by molecular dynamics (MD) simulations using explicit solvent. Thus a comparison of distance averages (〈r −6〉−1/6 or 〈r −3〉1/3) over the MD trajectory with NOE (or ROE) derived distances is no longer necessary, the (back)calculation of the complete spectra becomes possible. In the present study we use two 200 ns trajectories of a heptapeptide of β-amino acids in methanol at two different temperatures to obtain theoretical ROESY spectra by calculating the exact spectral densities for the interproton vectors and the full relaxation matrix. Those data are then compared with the experimental ones. This analysis permits to test some of the assumptions and approximations that generally have to be made to interpret NMR spectra, and to make a more reliable prediction of the conformational equilibrium that leads to the experimental spectrum.
Similar content being viewed by others
References
Boelens, R., Koning, T.M.G., van der Marel, G.A., van Boom, J.H. and Kaptein, R. (1989) J. Magn. Reson., 82, 290-308.
Brüschweiler, R. and Case, D. (1994) Progr. NMR Spectrosc., 26, 27-58.
Daura, X., Antes, I., van Gunsteren, W.F., Thiel, W. and Mark, A.E. (1999a) Proteins, 36, 542-555.
Daura, X., Jaun, B., Seebach, D., van Gunsteren, W.F. and Mark, A.E. (1998) J. Mol. Biol., 280, 925-932.
Daura, X., van Gunsteren, W.F. and Mark, A.E. (1999b) Proteins, 34, 269-280.
Edmondson, S.P. (1992) J. Magn. Reson., 98, 283-298.
Edmondson, S.P. (1994) J. Magn. Reson., B103, 222-233.
Fischer, M.W.F., Majumdar, A. and Zuiderweg, E.R.P. (1998) Progr. NMR Spectrosc., 33, 207-272.
Keepers, J.W. and James, T.L. (1984) J. Magn. Reson., 57, 404-426.
Lipari, G. and Szabo, A. (1982) J. Am. Chem. Soc., 104, 4546-4559.
Macura, S. and Ernst, R.R. (1980) Mol. Phys., 41, 95-117.
Neuhaus, D. and Williamson, M. (1989) The Nuclear Overhauser Effect in Structural and Conformational Analysis, VCH, New York, NY.
Olejniczak, E.T. (1989) J. Magn. Reson., 81, 392-394.
Seebach, D., Ciceri, P.E., Overhand, M., Jaun, B., Rigo, D., Oberer, L., Hommel, U., Amstutz, R. and Widmer, H. (1996) Helv. Chim. Acta, 79, 2043-2066.
Tropp, J. (1980) J. Chem. Phys., 72, 6035-6043.
Walser, R., Mark, A.E., van Gunsteren, W.F., Lauterbach, M. and Wipff, G. (2000) J. Chem. Phys., 112, 10450-10459.
Woessner, D.E. (1962) J. Chem. Phys., 36, 1-4.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Peter, C., Daura, X. & van Gunsteren, W.F. Calculation of NMR-relaxation parameters for flexible molecules from molecular dynamics simulations. J Biomol NMR 20, 297–310 (2001). https://doi.org/10.1023/A:1011241030461
Issue Date:
DOI: https://doi.org/10.1023/A:1011241030461