Abstract
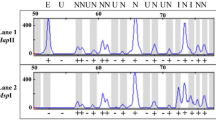
We investigated the CpG methylation status of the sequence CCGG in the rice genome by using methylation-sensitive AFLP and subsequent Southern analyses with the isolated AFLP fragments as probes. CpGs located in single- or low-copy-sequence regions could be grouped into two classes on the basis of their methylation status: methylation status at the class 1 CpG sites was conserved among genetically diverse rice cultivars, whereas cultivar-specific differential methylation was frequently detected among the cultivars at the class 2 CpG sites. The frequency of occurrence of methylation polymorphism between a pair of cultivars was not related to the genetic distance between the two. Through mapping, five class 2 CpG sites were localized on different chromosomes and were not clustered together in the genome. Segregation analysis of the cultivar-specific methylations with their target sites indicated that the differential methylation was stably inherited in a Mendelian fashion over 6 generations, although alterations in the methylation status at the class 2 CpG sites were observed with a low frequency.
Similar content being viewed by others
References
Ashikawa, I., Fukuta, Y., Tamura, K. and Yagi, T. 1999. Application of an AFLP technique that uses non-radioactive fluorescent primers to detection of genetic diversity in Japanese rice cultivars and cloning of DNA sequences derived from an indica genome. Breed. Sci. 49: 225–231.
Banks, J.A. and Fedoroff, N. 1989. Patterns of developmental and heritable change in methylation of the suppressor-mutator transposable element. Dev. Genet. 10: 425–437.
Bennetzen, J.L., Schrick, K., Springer, P.S., Brown, W.E. and San Miguel, P. 1994. Active maize genes are unmodified and flanked by diverse classes of modified, highly repetitive DNA. Genome 37: 565–576.
Constancia, M., Pickard, B., Kelsey, G. and Reik, W. 1998. Imprinting Mechanisms. Genome Res. 8: 881–900.
Cubas, P., Vincent, C. and Coen, E. 1999. An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401: 157–161.
Finnegan, E.J., Peacock, W.J. and Dennis, E.S. 1996. Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc. Natl. Acad. Sci. USA 93: 8449–8454.
Finnegan, E.J., Genger, R.K., Kovac, K., Peacock, W.J. and Dennis, E.S. 1998. DNA methylation and the promotion of flowering by vernalization. Proc. Natl. Acad. Sci. USA 95: 5824–5829.
Gardiner-Garden, M. and Frommer, M. 1987. CpG islands in vertebrate genomes. J. Mol. Biol. 196: 261–282.
Gruenbaum, Y., Naveh-Many, T., Cedar, H. and Razin, A. 1981. Sequence specificity of methylation in higher plant DNA. Nature 292: 860–862.
Jablonka, E., Goiten, R., Marcus, M. and Cedar, H. 1985. DNA hypomethylation causes an increase in DNase I sensitivity and advance in the timing of replication of the entire X chromosome. Chromosoma 93: 152–156.
Jacobsen, S.E. and Meyerowitz, E.M. 1997. Hypermethylated SUPERMAN epigenetic alleles in Arabidopsis. Science 277: 1100–1103.
Kakutani, T., Jeddeloh, J.A., Flowers, S.K., Munakata, K. and Richards, E. 1996. Developmental abnormalities and epimuta-tions associated with DNA hypomethylation mutations. Proc. Natl. Acad. Sci. USA 93: 12406–12411.
Kakutani, T., Munakata, K., Richards, E.J., Hirochika, H. 1999. Meiotically and mitotically stable inheritance of DNA hypomethylation induced by ddm1 mutation of Arabidopsis thaliana. Genetics 151: 831–838.
Lander, E.S., Green, P., Abrahamson, J., Barlow, A., Daly, M.J., Lincoln, S.E. and Newburg, lL. 1987. MAPMAKER: an interac-tive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181.a
Li, E., Bestor, T.H. and Jaenisch, R. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69: 915–926.
Lin, S.Y., Sasaki, T. and Yano, M. 1998. Mapping quantitative trait loci controlling seed dormancy and heading date in rice, Oryza sativa L., using backcross inbred lines. Theor. Appl. Genet. 96: 997–1003.
Lund, G., Messing, J. and Viotti, A. 1995. Endosperm-specific demethylation and activation of specific alleles of alpha-tubulin genes of Zea mays L. Mol. Gen. Genet. 246: 716–722.
Messeguer, R., Ganal, M.W., Steffens, J.C. and Tanksley, S.D. 1991. Characterization of the level, target sites and inheritance of cy-tosine methylation in tomato nuclear DNA. Plant. Mol. Biol. 16: 753–770.
Murray, M.G. and Thompson, W.F. 1980. Rapid isolation of high molecular weight plant DNA. Nucl. Acids Res. 8: 4321–4325.
Rabinowicz, P.D., Schutz, K., Dedhia N., Yordan, C., Parnell, L.D., Stein, L., McCombie, W.R. and Martienssen, R.A. 1999. Dif-ferential methylation of genes and retrotransposons facilitates shotgun sequencing of the maize genome. Nature Genet.23: 305–308.
Razin, A. 1998. CpG methylation, chromatin structure and gene silencing-a three-way connection. EMBO J. 17: 4905–4908.
Razin, A. and Cedar, H. 1991. DNA methylation and gene expression. Microbiol. Rev. 55: 451–458.
Reyna-Lopez, G.E., Simpson, J. and Ruiz-Herrera, J. 1997. Differences in DNA methylation patterns are detectable during the dimorphic transition of fungi by amplification restriction polymorphisms. Mol. Gen. Genet. 253: 703–710.
Richards, E.J. 1997. DNA methylation and plant development. Trends Genet. 13: 319–323.
Ronemus, M.J., Galbiati, M., Ticknor, C., Chen, J. and Dellaporta, S.L. 1996. Demethylation-induced developmental pleiotropy in Arabidopsis. Science 273: 654–657.
Rossi, V., Motto, M. and Pellegrini, L. 1997. Analysis of the methy-lation pattern of the maize opaque-2 (O2)promoterandin vitro binding studies indicate that the O2 B-Zip protein and other en-dosperm factors can bind to methylated target sequences. J. Biol. Chem. 272: 13758–13765.
Triglia, T., Peterson, M.G. and Kemp, D.J. 1988. A procedure for in vitro amplification of DNA segments that lie outside of the boundaries of known sequences. Nucl. Acids Res. 16: 8186.
Walker, E.L. 1998. Paramutation of the r1 locus of maize is as-sociated with increased cytosine methylation. Genetics 148: 1973–1981.
Xiong, L.Z., Xu, C.G., Saghai Maroof, M.A. and Zhang, Q. 1999. Patterns of cytosine methylation in an elite rice hybrid and its parental lines, detected by a methylation-sensitive amplification polymorphism technique. Mol. Gen. Genet. 261: 439–446.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Ashikawa, I. Surveying CpG methylation at 5′-CCGG in the genomes of rice cultivars. Plant Mol Biol 45, 31–39 (2001). https://doi.org/10.1023/A:1006457321781
Issue Date:
DOI: https://doi.org/10.1023/A:1006457321781