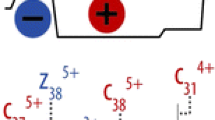
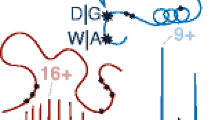
Abstract
Secondary fragmentations of three synthetic peptides (human αA crystallin peptide 1-11, the deamidated form of human βB2 crystallin peptide 4-14, and amyloid β peptide 25-35) were studied in both electron capture dissociation (ECD) and electron-transfer dissociation (ETD) mode. In ECD, in addition to c and z· ion formations, charge remote fragmentations (CRF) of z· ions were abundant, resulting in internal fragment formation or partial/entire side-chain losses from amino acids, sometimes several residues away from the backbone cleavage site, and to some extent multiple side-chain losses. The internal fragments were observed in peptides with basic residues located in the middle of the sequences, which was different from most tryptic peptides with basic residues located at the C-terminus. These secondary cleavages were initiated by hydrogen abstraction at the α-, β-, or γ-position of the amino acid side chain. In comparison, ETD generates fewer CRF fragments than ECD. This secondary cleavage study will facilitate ECD/ETD spectra interpretation, and help de novo sequencing and database searching.
Article PDF
Similar content being viewed by others
References
Masterson, D. S.; Yin, H. Y.; Chacon, A.; Hachey, D. L.; Norris, J. L.; Porter, N. A. Lysine Peroxycarbamates: Free Radical-Promoted Peptide Cleavage. J. Am. Chem. Soc. 2004, 126, 720–721.
Yin, H.; Chacon, A.; Porter, N. A.; Yin, H. Y.; Masterson, D. S. Free Radical-Induced Site-Specific Peptide Cleavage in the Gas Phase: Low-Energy Collision-Induced Dissociation in ESI- and MALDI Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2007, 18, 807–816.
Hodyss, R.; Cox, H. A.; Beauchamp, J. L. Bioconjugates for Tunable Peptide Fragmentation: Free Radical Initiated Peptide Sequencing (FRIPS). J. Am. Chem. Soc. 2005, 127, 12436–12437.
Hao, G.; Gross, S. S. Electrospray Tandem Mass Spectrometry Analysis of S- and N-Nitrosopeptides: Facile Loss of NO and Radical-Induced Fragmentation. J. Am. Soc. Mass Spectrom. 2006, 17, 1725–1730.
Wee, S.; Mortimer, A.; Moran, D.; Wright, A.; Barlow, C. K.; O’Hair, R. A. J.; Radom, L.; Easton, C. J. Gas-Phase Regiocontrolled Generation of Charged Amino Acid and Peptide Radicals. Chem. Commun. 2006, (40), 4233–4235.
Diedrich, J. K.; Julian, R. R. Site-Specific Radical Directed Dissociation of Peptides at Phosphorylated Residues. J. Am. Chem. Soc. 2008, 130, 12212–12213.
Ly, T.; Julian, R. R. Residue-Specific Radical-Directed Dissociation of Whole Proteins in the Gas Phase. J. Am. Chem. Soc. 2008, 130, 351–358.
Chu, I. K.; Rodriquez, C. F.; Lau, T. C.; Hopkinson, A. C.; Siu, K. W. M. Molecular Radical Cations of Oligopeptides. J. Phys. Chem. B. 2000, 104, 3393–3397.
Bagheri-Majdi, E.; Ke, Y. Y.; Orlova, G.; Chu, I. K.; Hopkinson, A. C.; Siu, K. W. M. Copper-Mediated Peptide Radical Ions in the Gas Phase. J. Phys. Chem. B. 2004, 108, 11170–11181.
Barlow, C. K.; McFadyen, W. D.; O’Hair, R. A. J. Formation of Cationic Peptide Radicals by Gas-Phase Redox Reactions with Trivalent Chromium, Manganese, Iron, and Cobalt Complexes. J. Am. Chem. Soc. 2005, 127, 6109–6115.
Laskin, J.; Yang, Z. B.; Lam, C.; Chu, I. K. Charge-Remote Fragmentation of Odd-Electron Peptide Ions. Anal. Chem. 2007, 79, 6607–6614.
Sun, Q. Y.; Nelson, H.; Ly, T.; Stoltz, B. M.; Julian, R. R. Side-Chain Chemistry Mediates Backbone Fragmentation in Hydrogen Deficient Peptide Radicals. J. Proteome Res. 2009, 8, 958–966.
Ly, T.; Yin, S.; Loo, J. A.; Julian, R. R. Electron-Induced Dissociation of Protonated Peptides Yields Backbone Fragmentation Consistent with a Hydrogen-Deficient Radical. Rapid Commun. Mass Spectrom. 2009, 23, 2099–2101.
Lioe, H.; O’Hair, R. A. J. Comparison of Collision-Induced Dissociation and Electron-Induced Dissociation of Singly Protonated Aromatic Amino Acids, Cystine, and Related Simple Peptides Using a Hybrid Linear Ion Trap-FT-ICR Mass Spectrometer. Anal. Bioanal. Chem. 2007, 389, 1429–1437.
Budnik, B. A.; Haselmann, K. F.; Elkin, Y. N.; Gorbach, V. I.; Zubarev, R. A. Applications of Electron-Ion Dissociation Reactions for Analysis of Polycationic Chitooligosaccharides in Fourier Transform Mass Spectrometry. Anal. Chem. 2003, 75, 5994–6001.
Zubarev, R. A.; Kelleher, N. L.; McLafferty, F. W. Electron Capture Dissociation of Multiply Charged Protein Cations: A Nonergodic Process. J. Am. Chem. Soc. 1998, 120, 3265–3266.
Zubarev, R. A.; Haselmann, K. F.; Budnik, B.; Kjeldsen, F.; Jensen, F. Towards an Understanding of the Mechanism of Electron-Capture Dissociation: A Historical Perspective and Modern Ideas. Eur. J. Mass Spectrom. 2002, 8, 337–349.
Syrstad, E. A.; Turecek, F. Toward a General Mechanism of Electron Capture Dissociation. J. Am. Soc. Mass Spectrom. 2005, 16, 208–224.
Syka, J. E. P.; Coon, J. J.; Schroeder, M. J.; Shabanowitz, J.; Hunt, D. F. Peptide and Protein Sequence Analysis by Electron Transfer Dissociation Mass Spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 9528–9533.
Mikesh, L. M.; Ueberheide, B.; Chi, A.; Coon, J. J.; Syka, J. E. P.; Shabanowitz, J.; Hunt, D. F. The Utility of ETD Mass Spectrometry in Proteomic Analysis. BBA-Prot. Proteom. 2006, 1764, 1811–1822.
Coon, J. J.; Shabanowitz, J.; Hunt, D. F.; Syka, J. E. P. Electron Transfer Dissociation of Peptide Anions. J. Am. Soc. Mass Spectrom. 2005, 16, 880–882.
Laskin, J.; Futrell, J. H.; Chu, I. K. Is Dissociation of Peptide Radical Cations an Ergodic Process? J. Am. Chem. Soc. 2007, 129, 9598–9599.
Zhang, L.; Reilly, J. P. Radical-Driven Dissociation of Odd-Electron Peptide Radical Ions Produced in 157 nm Photodissociation. J. Am. Soc. Mass Spectrom. 2009, 20, 1378–1390.
O’Connor, P. B.; Lin, C.; Cournoyer, J. J.; Pittman, J. L.; Belyayev, M.; Budnik, B. A. Long-Lived Electron Capture Dissociation Product Ions Experience Radical Migration Via Hydrogen Abstraction. J. Am. Soc. Mass Spectrom. 2006, 17, 576–585.
Lin, C.; O’Connor, P. B.; Cournoyer, J. J. Use of a Double Resonance Electron Capture Dissociation Experiment to Probe Fragment Intermediate Lifetimes. J. Am. Soc. Mass Spectrom. 2006, 17, 1605–1615.
Cooper, H. J.; Hudgins, R. R.; Hakansson, K.; Marshall, A. G. Characterization of Amino Acid Side-Chain losses in Electron Capture Dissociation. J. Am. Soc. Mass Spectrom. 2002, 13, 241–249.
Cooper, H. J.; Hudgins, R. R.; Hakansson, K.; Marshall, A. G. Secondary Fragmentation of Linear Peptides in Electron Capture Dissociation. Int. J. Mass Spectrom. 2003, 228, 723–728.
Fung, Y. M. E.; Chan, T. W. D. Experimental and Theoretical Investigations of the Loss of Amino Acid Side Chains in Electron Capture Dissociation of Model Peptides. J. Am. Soc. Mass Spectrom. 2005, 16, 1523–1535.
Savitski, M. M.; Nielsen, M. L.; Zubarev, R. A. Side-Chain Losses in Electron Capture Dissociation to Improve Peptide Identification. Anal. Chem. 2007, 79, 2296–2302.
Falth, M.; Savitski, M. M.; Nielsen, M. L.; Kjeldsen, F.; Andren, P. E.; Zubarev, R. A. Analytical Utility of Small Neutral Losses from Reduced Species in Electron Capture Dissociation Studied Using SwedECD Database. Anal. Chem. 2008, 80, 8089–8094.
Kjeldsen, F.; Zubarev, R. Secondary Losses Via γ-Lactam Formation in Hot Electron Capture Dissociation: A Missing Link to Complete De Novo Sequencing of Proteins? J. Am. Chem. Soc. 2003, 125, 6628–6629.
Haselmann, K. F.; Budnik, B. A.; Kjeldsen, F.; Polfer, N. C.; Zubarev, R. A. Can the (M·-X) Region in Electron Capture Dissociation Provide Reliable Information on Amino Acid Composition of Polypeptides? Eur. J. Mass Spectrom. 2002, 8, 461–469.
Cooper, H. J.; Hakansson, K.; Marshall, A. G.; Hudgins, R. R.; Haselmann, K. F.; Kjeldsen, F.; Budnik, B. A.; Polfer, N. C.; Zubarev, R. A. Letter: The Diagnostic Value of Amino Acid Side-Chain Losses in Electron Capture Dissociation of Polypeptides. Comment on: “Can the (M·-X) Region in Electron Capture Dissociation Provide Reliable Information on Amino Acid Composition of Polypeptides?” Eur. J. Mass Spectrom. 2002, 8, 461–469. Eur. J. Mass Spectrom. 2003, 9, 221–222.
Cheng, C.; Gross, M. L. Applications and Mechanisms of Charge-Remote Fragmentation. Mass Spectrom. Rev. 2000, 19, 398–420.
Gross, M. L. Charge-Remote Fragmentation: An Account of Research on Mechanisms and Applications. Int. J. Mass Spectrom. 2000, 200, 611–624.
Jensen, N. J.; Tomer, K. B.; Gross, M. L. Gas-Phase Ion Decompositions Occurring Remote to a Charge Site. J. Am. Chem. Soc. 1985, 107, 1863–1868.
Adams, J.; Gross, M. L. Energy Requirements for Remote Charge Site Ion Decompositions and Structural Information from Collisional Activation of Alkali-Metal Cationized Fatty Alcohols. J. Am. Chem. Soc. 1986, 108, 6915–6921.
Adams, J.; Gross, M. L. Charge-Remote Fragmentations of Closed-Shell Ions—a Thermolytic Analogy. J. Am. Chem. Soc. 1989, 111, 435–440.
Gross, M. L. Charge-Remote Fragmentations—Method, Mechanism, and Applications. Int. J. Mass Spectrom. Ion Process. 1992, 118, 137–165.
Han, H. L.; Xia, Y.; McLuckey, S. A. Ion Trap Collisional Activation of c and z· Ions Formed Via Gas-Phase Ion/Ion Electron Transfer Dissociation. J. Proteome Res. 2007, 6, 3062–3069.
Leymarie, N.; Costello, C. E.; O’Connor, P. B. Electron Capture Dissociation Initiates a Free Radical Reaction Cascade. J. Am. Chem. Soc. 2003, 125, 8949–8958.
Jebanathirajah, J. A.; Pittman, J. L.; Thomson, B. A.; Budnik, B. A.; Kaur, P.; Rape, M.; Kirschner, M.; Costello, C. E.; O’Connor, P. B. Characterization of a New qQq-FTICR Mass Spectrometer for Post-Translational Modification Analysis and Top-Down Tandem Mass Spectrometry of Whole Proteins. J. Am. Soc. Mass Spectrom. 2005, 16, 1985–1999.
O’Connor, P. B.; Pittman, J. L.; Thomson, B. A.; Budnik, B. A.; Cournoyer, J. C.; Jebanathirajah, J.; Lin, C.; Moyer, S.; Zhao, C. A New Hybrid Electrospray Fourier Transform Mass Spectrometer: Design and Performance Characteristics. Rapid Commun. Mass Spectrom. 2006, 20, 259–266.
Swaney, D. L.; McAlister, G. C.; Wirtala, M.; Schwartz, J. C.; Syka, J. E. P.; Coon, J. J. Supplemental Activation Method for High-Efficiency Electron-Transfer Dissociation of Doubly Protonated Peptide Precursors. Anal. Chem. 2007, 79, 477–485.
Cooper, H. J. Investigation of the Presence of b Ions in Electron Capture Dissociation Mass Spectra. J. Am. Soc. Mass Spectrom. 2005, 16, 1932–1940.
Liu, H. C.; Hakansson, K. Abundant b-Type Ions Produced in Electron Capture Dissociation of Peptides Without Basic Amino Acid Residues. J. Am. Soc. Mass Spectrom. 2007, 18, 2007–2013.
Li, X.; Cournoyer, J. J.; Lin, C.; O’Connor, P. B. The Effect of Fixed Charge Modifications on Electron Capture Dissociation. J. Am. Soc. Mass Spectrom. 2008, 19, 1514–1526.
Belyayev, M. A.; Cournoyer, J. J.; Lin, C.; O’Connor, P. B. The Effect of Radical Trap Moieties on Electron Capture Dissociation Spectra of Substance P. J. Am. Soc. Mass Spectrom. 2006, 17, 1428–1436.
Kjeldsen, F.; Haselmann, K. F.; Budnik, B. A.; Jensen, F.; Zubarev, R. A. Dissociative Capture of Hot (3–13 eV) Electrons by Polypeptide Polycations: An Efficient Process Accompanied by Secondary Fragmentation. Chem. Phys. Lett. 2002, 356, 201–206.
Cournoyer, J. J.; Pittman, J. L.; Ivleva, V. B.; Fallows, E.; Waskell, L.; Costello, C. E.; O’Connor, P. B. Deamidation: Differentiation of Aspartyl from Isoaspartyl Products in Peptides by Electron Capture Dissociation. Protein Sci. 2005, 14, 452–463.
Cournoyer, J. J.; Lin, C.; O’Connor, P. B. Detecting Deamidation Products in Proteins by Electron Capture Dissociation. Anal. Chem. 2006, 78, 1264–1271.
Cournoyer, J. J.; Lin, C.; Bowman, M. J.; O’Connor, P. B. Quantitating the Relative Abundance of Isoaspartyl Residues in Deamidated Proteins by Electron Capture Dissociation. J. Am. Soc. Mass Spectrom. 2007, 18, 48–56.
Li, X.; Cournoyer, J. J.; Lin, C.; O’Connor, P. B. Use of O-18 Labels to Monitor Deamidation During Protein and Peptide Sample Processing. J. Am. Soc. Mass Spectrom. 2008, 19, 855–864.
O’Connor, P. B.; Cournoyer, J. J.; Pitteri, S. J.; Chrisman, P. A.; McLuckey, S. A. Differentiation of Aspartic and Isoaspartic Acids Using Electron Transfer Dissociation. J. Am. Soc. Mass Spectrom. 2006, 17, 15–19.
Kjeldsen, F.; Haselmann, K. F.; Sorensen, E. S.; Zubarev, R. A. Distinguishing of Ile/Leu Amino Acid Residues in the PP3 Protein by (Hot) Electron Capture Dissociation in Fourier Transform Ion Cyclotron Resonance Mass Spectrometry. Anal. Chem. 2003, 75, 1267–1274.
Savitski, M. M.; Kjeldsen, F.; Nielsen, M. L.; Zubarev, R. A. Hydrogen Rearrangement to and from Radical z Fragments in Electron Capture Dissociation of Peptides. J. Am. Soc. Mass Spectrom. 2007, 18, 113–120.
Lin, C.; Cournoyer, J. C.; O’Connor, P. B. Probing the Gas Phase Folding Kinetics of Peptide Ions by IR Activated DR-ECD. J. Am. Soc. Mass Spectrom. 2008, 19, 780–789.
Author information
Authors and Affiliations
Corresponding author
Additional information
Published online January 18, 2010
Rights and permissions
About this article
Cite this article
Li, X., Lin, C., Han, L. et al. Charge remote fragmentation in electron capture and electron transfer dissociation. J Am Soc Mass Spectrom 21, 646–656 (2010). https://doi.org/10.1016/j.jasms.2010.01.001
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1016/j.jasms.2010.01.001