Abstract
In the era of predicted significant environmental change, genetic conservation and monitoring of long-lived forest trees are of paramount importance for the survival of natural populations. Genetic conservation aims to protect and preserve genetic variation, vital for the maintenance of adaptive potential within populations and species. This paper discusses the advances made in gene conservation and genetic monitoring approaches in particular that aim to put into practise a concept that was developed more than 25 years ago, but rarely put into practise. Genetic monitoring, the quantification of temporal changes in population genetic variation and structure, elucidates processes that maintain genetic variation in natural populations, introduces prognosis and helps define tools for the management of forest genetic resources. Based on the geneecological approach, proposed indicators reflect the assessment of genetic variation, genetic drift, gene flow (mating system) and natural selection. Indicators are evaluated in contemporary monitoring schemes by 7–11 verifiers. Genomic data are expected to increase the precision of estimates of adaptive genetic potential and of population genetic parameters due to the significantly higher number of markers assayed. The transition from genetic to genomic monitoring should provide an enhanced potential for disentangling natural selection from demography, and deciphering the association of genetic variation to environmental gradients. In the future, genetic monitoring will be more limited by time-consuming procedures and funding constraints in the assessment of demographic parameters, as well as by conceptual and analytical weaknesses of biostatistical tools, than by genotyping needs and the amount of available sequence data. Its important contribution to applied forest management can be foreseen.
Similar content being viewed by others
Introduction
In the past 300 years, the world has lost more than 700 million ha of tropical forest and more than 300 million ha of temperate forest, mainly due to anthropogenic impacts. Large forest destruction started during the Holocene and continued to the present day. In ancient Greece for instance, van Andel et al. [1] found repeated episodes of deforestation followed by soil erosion since the late Neolithic period. It is estimated that about 45 % of world forests have been lost, and about 90 % of this loss has occurred since the start of the twentieth century [2]. Continual forest destruction has a profound effect on biodiversity loss. Today, the rate of population extinction and species loss may be 1000× higher than background levels [2].
Despite these losses, trees still constitute more than 80 % of continental biomass and forests harbour more than 50 % of terrestrial biodiversity. Forests continue to offer a sustainable source of primary production that includes wood, paper, tree-crop foods, pharmaceuticals, bioenergy, as well as a multitude of ecological services. Forests preserve biodiversity and the genetic heritage of natural ecosystems and play a key role in carbon sequestration and storage, climate regulation, production and preservation of water quality, soil protection and erosion control. A large number of trees are keystone species for many natural ecosystems and form a cornerstone for the support of different biological communities within the natural environment [3].
Given the role of forest ecosystems in sustaining life on earth, the genetic resources and biodiversity maintained in forest systems constitute intergenerational resources of immense social, economic and environmental importance. The recently published State of the World’s Forest Genetic Resources FAO [4] indicates that about 4000 valuable tree species are considered to be threatened in some way. Almost 2300 tree species have been listed as national priorities for conservation and management [5]. Indeed, to counteract species extinction and biodiversity loss, the concept of biological conservation has been advanced. This refers to the science and management activities which ensure the survival of the maximum diversity of species and the maintenance of genetic variation within species. Since 1993, the United Nation’s Convention on Biological Diversity (CBD) [6] has been the most important international political instrument dealing with issues of biodiversity conservation and reduction of biodiversity loss. The conservation of forest genetic resource refers to the science, management and policy activities that ensure their continued availability and existence. It is based on two strategies: in situ (“in place”) conservation implies the continuing maintenance of a population within the environment where it originally evolved and to which it is assumed to be adapted, and ex situ (“out of place”) conservation serves to capture and maintain genetic variation in plantations, gene or seed banks, as well as tissue or DNA sample collections. In this paper, the term “conservation” refers to dynamic in situ gene conservation, i.e. the long-term conservation of evolutionary processes within tree populations in order to maintain their adaptive potential in their original environment.
The need for global targets for reducing biodiversity loss and the means to monitor progress towards those goals led to the development of global biodiversity indicators [7••]. CBD [6] and several other international (e.g. Montreal Process, ITTO) and regional processes (e.g. Helsinki Process) have called for the establishment of criteria and indicators for genetic diversity [8]. However, until recently, there has been an almost worldwide lack of international or national interest regarding the utilization of genetic indicators in conservation policies [9]. Recently, discussion has intensified on using genetic monitoring for assessing the status of forest gene conservation. Genetic monitoring is the quantification of temporal changes in population genetic variation and structure [10••]. Contemporary major issues focus on theoretical developments regarding the application of an efficient genetic monitoring system in practice [10••] and on the development of genetic monitoring applications at different levels: local (such as the application of forest genetic monitoring in Germany [11•]), regional (such as the project “LIFE for European Forest Genetic Monitoring System; http://www.lifegenmon.si/) and continental (such as the development of Pan-European system for the genetic monitoring of forest genetic resources by the European Forest Genetic Resources Programme, EUFORGEN; http://www.euforgen.org/[12••]).
The aim of this paper is to contribute to the discussion on the past, present and future of gene conservation and genetic monitoring of temperate forests. It attempts to evaluate the scientific basis for gene conservation and genetic monitoring developed thus far discuss their current application and their future, especially at the transition towards the genomic era, i.e. consider the development of genomic conservation and genomic monitoring of forest genetic resources.
Conservation Genetics of Forest Tree Populations
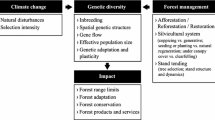
There are two major issues that are driving the need for immediate, extensive and comprehensive conservation of forest genetic resources: (a) the vast and fast climatic changes and (b) the direct adverse anthropogenic influence and activity. Both can be loosely encompassed under the term environmental change. Global temperature is expected to rise about 1.8–4.0 °C during the twenty-first century [13] with unknown effects on genotype × environment interactions. Climate warming may induce an estimated shift of species spatial distributions more than 6 km towards the poles and 1 m in elevation, per decade [14]. Such alterations may result in population spatial shifts, fragmentation, reduction of population size or even extinction in mountainous ranges as a result of the “summit trap” phenomenon. Environmental change affects genetic diversity at various scales in a diverse, complex and unpredictable manner that includes alternations of natural distribution envelopes, changes in species interactions and phenology, and reduction of genetic diversity and adaptive evolutionary potential. Even when environmental variation does not result in variation in species and population demography and growth rate, many studies identify interaction between environmental variation and population density in several species [15]. Population density often reflects census population size (a proxy of effective population size, N e [16]) and mean individual fitness [17] and influences spatial genetic structure and the mating system (preference for selfing or outcrossing). Also, population density may be an indicator of habitat quality, marginality and fragmentation. The development of forest conservation policy during the twenty-first century presents several challenges [18•]. The issue of implementing genetic monitoring and identifying suitable indicators and verifiers to do so is one of the most important among them.
The Central Dogma of Conservation Genetics
The central dogma of conservation genetics states that genetic variability is beneficial, hence worth preserving to the greatest extent [15]. As genetic variation is important in maintaining the adaptive potential of species/populations and the fitness of individuals, there is a general consensus that conservation of biodiversity ultimately depends on the conservation of genetic diversity and that increasing genetic variance enhances the probability of population survival [19].
Heritable genetic variation is a prerequisite for adaptation and evolution, and therefore, an essential aim of conservation genetics is to quantify the amount of heritable genetic variation present. Theory predicts that populations will only persist if the rate of adaptive evolution at least matches the rate of environmental change. The phenotypic response to selection is the product of additive genetic variance (σΑ 2), which describes the cumulative effect of the individual genes, times the selection differential (s) or the standardized intensity (i) of the selection coefficient [20]. In a population, the actual degree of adaptation is the residual effect of the dynamic interaction between the selective pressure acting on the population and gene flow. Hence, high levels of gene flow may introduce essential new genes for future adaptation and increase the capacity to adapt or can reduce/impede the capacity of adaptation to local conditions [21, 22].
The latter possibility has led to a criticism of the central dogma: in a constant environment, genetic variance in a quantitative trait creates in each generation a segregational load due to stabilizing selection against individuals that deviate from the optimum phenotype [15]. Hence, genetic variability cannot be regarded as “good” per se: it can be either beneficial or detrimental, depending on the pattern of environmental change [23]. This challenge to the central dogma has some important implications for conservation and genetic monitoring. The assessment of genetic variation per se may not be enough for determining conservation goals and status. Gene flow and selection play an important role as well and they should also be assessed in conjunction to the wealth and the structure of genetic variation.
Inferring Conservation Need and Conservation Status
Determining the necessity for conservation, prioritizing conservation activities and managing forest genetic resources depend on evaluating the evolutionary potential of a population and its ability to adapt to changing environmental conditions. This assessment is not a once-for-all static process. Environmental change calls for dynamic continuous monitoring that preferably should have a temporal (ideally periodic) nature. In the forest, conservation primarily involves protected areas and delegated populations. Ideally, their assessment should be based on a series of demographic and genetic parameters, derived from markers and quantitative traits [24]. However, many demographic parameters (e.g. flowering) are difficult to assess regularly at a large scale. Furthermore, it is hardly possible to precisely assess the importance of variation of an adaptive trait for future conditions [25], while trait stability during ontogeny is usually not well known. Moreover, although quantitative genetics information is an invaluable resource [26], provenance–progeny tests are only available for a few species and are generally limited in terms of representation of the species natural distribution and/or protected areas, making this approach of limited practical value. Genetic markers offer a solution, mainly due to their recent explosion in numbers, types, facilitation of assessment and reduction in cost [3, 12••, 27]. The interpretation of variation patterns deduced from different genetic markers is not straightforward ([28], but see also discussion in the next paragraph). However, marker assessment in the long run will be far more economical than the evaluation of adaptive traits.
In successive monitoring assessment, demographic considerations may be essential to gene conservation in the shorter term [29]; nevertheless, in the longer term, population genetic parameters may be more important [25]. Thus, the best choice for the assessment of conservation need and status should be based on a combination of demographic and genetic parameters. The former would involve estimation of population density or census population size and an assessment of the phenotype (such as age, diameter and height), as well as an estimate of reproductive fitness (such as fructification, seed quality and regeneration). Genetic markers should be used to estimate population parameters including genetic diversity (i.e. allelic richness, expected heterozygosity, latent genetic potential, the latter being the difference between the total and the effective number of alleles summed over all loci [30]), gene flow (i.e. outcrossing and inbreeding rate, outcoming gene flow) and effective population size (N e ). N e is particularly important because when N e is small, genetic drift becomes much more important than selection [31]. Marker-based assessments may act, at least to some level, as surrogates for levels of adaptive variation. Molecular and quantitative trait variations have been theoretically correlated, although empirical studies indicated that their relationship is complex [32]. Mittell et al. [33] using data from 85 species (7166 quantitative and molecular genetic estimates from 412 studies) obtained a value of 26 % for the proportion of variance in quantitative genetic variation explained by molecular diversity. Neutral genetic variation may not be directly linked to quantitative genetic traits, but it may be significantly correlated to fitness [31, 32]. Meta-analyses suggest that natural selection has a predominant role as a cause of differentiation in quantitative traits, and neutral marker differentiation could at least be roughly predictive of genetic differentiation in quantitative traits [34]. Furthermore, there is extensive accumulating evidence linking the loss of marker heterozygosity to deleterious effects on population fitness [33, 35]. Especially in small populations where most of the fitness variation is neutral as genetic drift is predominant [36], genetic markers can be highly effective as a surrogate for fitness.
A Conservation Geneticist’s Dilemma: Small or Large Populations, Rare or Common Alleles?
In theory, the answer to the questions posed in the chapter title should be “both”. Nevertheless, we live in a world of limited resources, of limited time and of limited human power. Hence, genetic conservation falls within a large group of endeavours where proper prioritization is paramount. It is unquestionable that population size is very important in conservation (including the estimation of N e ). However, a number of studies suggest that only large populations experiencing relatively modest environmental change are likely to be rescued by evolution, since in small populations, genetic drift will mask selective pressures rendering most traits and genes effectively selectively neutral [15]. In that case, population adaptive potential is diminished, despite the presence of genetic variability. Small fragmented populations are frequently genetically depauperate and may suffer from inbreeding [37]. The combined effect of drift and inbreeding will augment genetic stochasticity; therefore, under changing environmental conditions, their long-term evolution and adaptation are questionable. Gene flow on the other hand can be a force for maintaining genetic diversity and alleviating inbreeding depression. Not surprisingly, increasing gene flow (or artificially introducing genes by reforestation, etc.) is a common rescue strategy in conservation. The presence or the deliberate introduction of gene flow is however risky: high levels of gene flow may cause outbreeding depression by introducing potentially maladapted genes which in turn may reduce the adaptive capability of local populations [38, 39]. Consequently, the conservation of small populations is a tricky issue.
Genetic conservation at the species level should focus, as a priority, on genetically distinct populations of appropriate N e . Genetic monitoring should be used as a tool in the assessment of their conservation status. Nevertheless, genetic monitoring should be immediately applied in the conservation of peculiar small populations (such as those of potential adaptive significance, for instance those growing at ecological or geographical margins).
The conservation of rare alleles is another conservation concern. Intuitively, conserving rare alleles comes to mind as a priority, as an extension of our approach towards rare species. Nevertheless, the phenotypic response to selection depends on additive genetic variance (σΑ 2), which in turn depends on gene frequencies. Rare and high-frequency alleles do not contribute considerably to additive genetic variance, which in fact is rapidly lost in small populations. In order for additive variance to be the fuel of adaptation, effective population sizes should be at least N e > 50. Evidently very low (or very high) frequency alleles may not be of primary concern for genetic monitoring.
Conservation Genomics of Forest Tree Populations
Genomics offers an unprecedented level of resolution in population genetic studies; it is currently feasible to substantially increase the numbers of populations, individuals per population and loci per individual studied at a fraction of earlier experimental costs. Conservation genomics has the goals of evaluating functional genomic variation in relation to drift and inbreeding and studying the mechanisms that relate low genetic variation to low fitness [39]. Conservation genomic studies in trees are still scarce, in spite of a universal acceptance of their value and potential [3, 12••, 27, 38–41] and despite reports indicating the vulnerability of in situ forest conservation due to climatic change [42]. Conservation genomics can potentially overcome the inherent limitations of common garden experiments by complementing provenance results with landscape genomic analysis and improve the estimation of adaptive genetic potential [43]. Genomics offers high precision estimates of genetic and demographic parameters and could result in high-resolution characterization of adaptive genetic variation in natura [44, 45•, 46••, 47]. A more informative delineation of conservation areas could be achieved since next-generation sequencing data will be more powerful and accurate, especially in cases where significant adaptive differentiation is expected among evolutionary significant units considered as candidates for gene conservation [38]. Measures of nucleotide diversity and divergence may be readily estimated [45•]. Departures from neutrality can be tested, and identification of allele-specific effects for thousands of genes through association mapping can be achieved. Typical forest genetic conservation studies used about 10 variable marker loci, but the analytical power of conservation genomics involves tens of thousands of loci [27]. Genomic analysis may determine which parts of the genome are responsible for local adaptation and therefore important to conserve. Genomic tools can investigate the interplay between population size (especially low N e ), genetic diversity and adaptation. In conclusion, genomics may prove to be of considerable value in the study of the genetic basis of local adaptation and its genetic architecture, even in non-model tree species [46••, 47]. The ability to evaluate both genetic and environmental threats will therefore lead to integrated conservation genomics. Thus, conservation of both neutral and adaptive genetic diversity may finally be tuned [27].
Genetic Monitoring of Forest Tree Populations
Objectives, Initial Approaches and Applications
The objective of genetic monitoring is to assess the current status of genetic resources and quantify relevant changes at a temporal scale in light of preserving long-term adaptive evolutionary potential. Genetic monitoring encompasses something more than a study method: by observing temporal changes in populations, causal components can be inferred and their relative importance can be evaluated. Genetic monitoring is an early detection mechanism that leads to management decisions aimed to lessen potential harmful effects before irreversible damage occurs. Thus, genetic monitoring is a prognostic tool and forms a method to secure the conservation of processes that maintain genetic variation in natural populations [10••].
In spite of the CBD [6] position that the measurement of genetic diversity trends is a crucial component in biodiversity assessment, until 2010, there has been an almost worldwide lack of international or national efforts regarding genetic monitoring [9]. Genetic monitoring was proposed more than 25 years ago [48], but it has only recently received attention [49, 50], mainly in fish (e.g. [51, 52]) and mammals (e.g. [53, 54]). Forest genetic monitoring was initially suggested by Namkoong et al. [24]. National programmes have only recently been proposed to conduct forest genetic monitoring either directly or indirectly (e.g. [11•, 54]). After evaluating the current status of the EUFORGEN conservation network, Schueler et al. [42] emphasized that there is a pressing need to intensify monitoring efforts and identify the most vulnerable gene conservation units. Evaluation schemes based on criteria, indicators and verifiers have been proposed. However, there is a scarcity of applied forest genetic monitoring studies, which is attributed to the complexity of the schemes proposed. This pertains to the use of several indicators within a single scheme, the difficulty in their assessment (time consuming, technical expertise, financial expense), the ambiguity in the assessment approach and the potential need of complex metadata (such as field trial data). Proposed schemes have considered up to 13 indicators and up to 23 verifiers for genetic monitoring, i.e. numbers that are rather unrealistic for regular application [10••]. The study which tested the German forest genetic monitoring system [11•] is an important example. This study confirmed that the use of multiple indicators does not always provide a clear conclusion as to the functionality of the genetic system of a tree population. It also confirmed the necessity of a genetic monitoring system, as problems in the genetic processes (selection, mating system, migration) may not be directly visible [11•]. As this was the first practical implementation of a genetic monitoring approach, questions pertaining to which indicators are needed, which may not be necessary in retrospect and if they deserve the same weighting, will remain at least until the follow-up assessment.
Theoretical Developments and Concept Evolution
Aravanopoulos [10••] attempted to use theoretical methods to formalize the forest genetic monitoring concept and approach. The paper emphasized that genetic monitoring is paramount for forest tree populations as adaptive responses to environmental change are better examined by a temporal approach, than by estimating response founded on the distribution of adaptive variation across populations in different environments. It proposed that genetic monitoring should have a solid scientific basis, a universal application (species-wide), set priorities for monitoring-conservation and use a system of criteria, indicators and verifiers that would extract the maximum amount of genetic information with a minimum number of parameters, including sample size and number of loci, as well as critical difference levels during temporal assessments that would warrant action [10••]. A criterion reflects a comprehensive objective; indicators are estimated periodically to reveal the direction of change in order to track progress in relation to the criterion; and verifiers are the metrics used to measure an indicator [7••, 55].
Aravanopoulos [10••] employed one criterion: the conservation of genetic diversity and adaptive evolutionary potential in natural populations. The application of genetic monitoring was prioritized as follows: (a) keystone species of biological and economical importance, starting from ecologically dominant species (aiming primarily at prevention), and (b) rare/endangered species (aiming primarily at restoration). Small populations (N e < 50) and rare alleles were not considered as special cases; however, populations at the rear edge of species distributions should, as they are generally expected to substantially suffer the consequences of climate change [10••, 18•, 56]. Moreover, it was proposed that genetic monitoring should concentrate on gene conservation units of selected species in a dynamic gene conservation scheme. The concept of dynamic gene conservation emphasizes maintenance of evolutionary processes within perennial plant populations to safeguard their potential for continuous adaptation. A gene conservation unit (GCU) refers to a population of a particular geographical location that has been selected for representing a high potential for adaptive evolution through the maintenance of population and metapopulation stability [57]. The proposed system is based on the assumption that within-species gene conservation units represent a series of differentiated metapopulations [10••].
The scientific basis for genetic monitoring has been proposed to be the geneecological approach: three factors are the major forces of evolution at the microscale: natural selection, genetic drift and gene flow. The effects of natural selection can lead to differentiation associated with local adaptation, while genetic drift can lead to differentiation associated with stochastic changes and genetic erosion. These are mediated by the action of gene flow that can lead to genetic homogenization. The action of mutation and other evolutionary forces can be considered as negligible for relatively short-term processes. This approach has also been implied in earlier works (e.g. [8, 24, 58]). The indicators proposed are directly reflecting the geneecological approach: (1) natural selection, (2) genetic drift and (3) gene flow [10••]. Seven verifiers have proposed to assess these indicators [10••], presented in Table 1. Therefore, this system is founded on the estimation of a minimum set of highly informative demographic and genetic parameters. Based on population genetics theory, Aravanopoulos [10••] proposed minimum sample sizes, types and numbers of marker loci, frequency of monitoring, as well as critical difference levels and thresholds between verifier measurements during temporal assessments.
More recently, in the preparation of the FAO’s State of the World’s Forest Genetic Resources Report [4], a thematic study on the genetic diversity indicators of evolutionary potential in tree species within and outside forests from the global to the local level was developed and summarized in a review paper [4, 7••]. Four types of indicators were proposed (state, pressure, response, benefit). Clearly, state indicators (indicators that refer to the condition and status of aspects of biodiversity) can be used as indicators for genetic monitoring at the local level. State operational indicator “Trends in population condition” (which can be regarded as equivalent to the “criterion” of genetic monitoring) is evaluated by two verifiable indicators: (1) demographic condition of selected populations (diversity in adaptive traits/genes) and (2) genetic condition of selected populations [7••]. These indicators are associated with 11 verifiers (Table 1). Out of the 11 verifiers proposed for assessing the evolutionary potential in tree species at the local level, seven are the same as those proposed by Aravanopoulos [10••] for genetic monitoring. Furthermore, indicator “demographic condition of selected populations” essentially includes the investigation of selection (i.e. indicator 1 of Aravanopoulos [10••]), while indicator “genetic condition of selected populations” essentially investigates genetic drift and gene flow (i.e. indicators 2 and 3 of Aravanopoulos [10••]).
In Europe, the development of a genetic monitoring system to assist the conservation of forest genetic resources has been discussed in EUFORGEN during phase III (2005–2009) and more intensively in phase IV (2010–2014) with the formulation of the Genetic Monitoring Working Group. The Working Group undertook the task to review genetic monitoring methods and to propose options for creating a Pan-European genetic monitoring system for the dynamic gene conservation of forest trees. This task has recently been completed [12••].
The Genetic Monitoring Working Group reflected on the threats to forest genetic resources and on the purpose of genetic monitoring as a means to address these threats. It discussed the definition, importance and historical development of genetic monitoring and considered the relevant terminology and indicator types. The Working Group addressed the main approaches in genetic monitoring, the relevant databases, DNA Repository Centres and eLabs. The main part of the Working Group Report describes the approach proposed for forest genetic monitoring at the Pan-European level. The approach is further exemplified in the proposed options for indicator and verifier assessment and in the approaches for identifying potential monitoring regions. Criteria for the selection of monitoring regions, principles and processes for selecting genetic monitoring units within monitoring regions and number of units per region were analysed and set. The Working Group has also pondered the design of genetic monitoring plots and sampling considerations. Last but not least, the cost of genetic monitoring was estimated taking into account a multitude of relevant parameters. The Working Group Report ends with conclusions and specific recommendations regarding the application of a forest genetic monitoring system in Europe [12••].
In particular, it suggested that genetic monitoring should start from the gene conservation units of selected species advanced in the Pan-European dynamic gene conservation system. It endorsed the use of the geneecological approach and the model of selected indicators and verifiers for population assessment. The proposed system includes only two indicators: (1) selection, and (2) genetic variation and mating system. Associated with these indicators is a set of nine verifiers (or up to 11 in the case of hybridization occurrence and in the case of dioecious species) in particular (Table 1):
-
for indicator “selection”: (1a) age/size class distribution, (1b) mortality, (1c) regeneration abundance, (1d) fructification and (1e) reproductive fitness in mast years
-
for indicator “genetic variation and mating system”: (2a) effective population size (N E ), (2b) allele/genotype frequencies, (2c) genetic diversity and (2d) outcrossing or actual inbreeding rate. In addition, for situations of hybridization (2e) interspecific hybridization percentage, and for dioecious species, (2f) sex ratio.
This scheme is concordant with that of Aravanopoulos [10••]; the alteration being the merging of indicators “genetic drift” and “gene flow” to one “genetic variation and mating system”. The verifiers proposed by Aravanopoulos [10••] are included in this system. Therefore, the EUFORGEN system is also a “tight” scheme in terms of the numbers of indicators and verifiers proposed, likewise to Aravanopoulos [10••] and Graudal et al. [7••]. These approaches of forest genetic monitoring include two to three indicators and 7–11 verifiers; they all are therefore systems that can realistically be applied in practice.
Lately, a European Union funding instrument for the environment and climate action, the LIFE Programme ( http://ec.europa.eu/environment/life/funding/lifeplus.htm), funded the project “LIFE for European Forest Genetic Monitoring System (LIFEGENMON; www.lifegenmon.si)”. This project is essentially a “proof of principle” application of forest genetic monitoring systems, as they have been theoretically proposed by Aravanopoulos [10••] and put in practise by Konnert et al. [11•]. The project implements genetic monitoring using the indicator and verifier scheme at the regional scale, namely on a transect spanning from Bavaria, Germany, to Mt. Olympus, Greece. It is expected that guidelines, an implementation manual and a decision support system for forest genetic monitoring users will be produced, based on genetic monitoring results as well as on practical findings considering the monitoring scheme in seven tree species: Abies alba/Abies borisii-regis complex, Fagus sylvatica, Fraxinus excelsior, Pinus nigra, Populus nigra, Prunus avium, and Quercus petraea/robur complex. It is expected that genetic monitoring results will be used in adaptive forest management to mitigate climate change threats for managed forest resources. Forest genetic monitoring can become a crucial component of any sustainable forest management system giving the possibility to detect potentially harmful changes of forest adaptability before they are seen at higher levels.
Genomic Monitoring of Forest Tree Populations
An understanding of forest tree population differentiation in terms of their adaptive potential is paramount for their gene conservation and monitoring. Forest population genetic differentiation, primarily from adaptive responses to local environmental changes, occurred since the last glacial period [45•]. For instance, trees frequently show geographical patterns of adaptive genetic variation, but neither field experiments nor molecular markers have fully disentangled the genetic basis of adaptation in trees ([3] and references therein).
The advent of genomics, in particular the availability of genome-wide sequences for non-model species, permits the simultaneous study of the effects of demographic history, migration and selection. The power of genomics lies in the ability to combine information on phenotype, genotype and local environment of large numbers of spatially referenced samples collected across landscape scales [43]. Genome-wide neutral loci are similarly affected by the demographic and evolutionary history of populations, while loci under selection will often behave differently and reveal “outlier” patterns of variation [59]. Hence, the association between environments and genotypes can be used in order to identify potential loci affected by selection. The meticulous evaluation of natural selection and demographic effects, the potential for their discrimination and the association with environmental gradients increase manifold the precision of the estimation of adaptive evolutionary potential.
An interesting example has been presented by Cullingham et al. [60] who studied the adaptive potential of lodgepole (Pinus contorta var. latifolia) and jack pine (Pinus banksiana), two species that have been exposed to different evolutionary pressures and their distributions are defined by dissimilar environments. They applied two approaches to detect intra- and interspecific adaptive variation using genomic data, the outlier approach (based on the assumption that loci under selection will have greater or lower variance than neutral loci, suggesting either divergent or stabilizing selection) and the spatial correlations of allele frequencies with environmental variable approach (environmental correlations will occur when the allele frequency at a locus is influenced by the environment and is therefore under environmental selection). The authors identified signatures of selection in 34 candidate outlier loci in lodgepole pine, 25 in jack pine and 43 in interspecific loci. Many of the outlier loci detected were correlated with environmental variation. Interestingly, the authors did not find any loci in common (within-species) between lodgepole and jack pine, a likely result of the species contrasting environmental adaptations. The number of consensus outliers detected ranged from 1 to 4 % and translates into thousands of adaptive sites. Moreover, environmental and spatial heterogeneity explained a large part of the variation in genetic diversity and contributed significantly to explaining allele variation among populations. This study illustrated the potential power of applying genomic approaches in monitoring. The estimation of genetic diversity and its relation to local environments and especially the detection of selection signals and environmental adaptation set a baseline that future assessments can refer to.
Genomic monitoring can therefore be defined as the quantification of temporal changes in population genomic variation and structure in the aim of the robust assessment of adaptive evolutionary potential. Genomic monitoring moves monitoring to the whole-genome level. In genomic monitoring, marker loci (mostly single nucleotide polymorphisms (SNPs) originating from candidate genes or from whole-genome scans) can be genotyped in trees from different target populations, such as gene conservation units, and tested for patterns of variation that signal selection. The detection of loci with unusually high or low levels of variation and differentiation (outlier loci) than putatively neutral ones is a powerful method to find loci under selection and to separate genome-wide effects that are caused by demographic processes from adaptive locus-specific effects [3, 61, 62]. Early detection of population declines can be facilitated by the use of SNPs in the estimation of N e [63]. Genomic monitoring will continue to use demographic parameters [10••, 12••], but will complement these with population genomic analysis. Both are needed for a proper delineation and assessment of gene conservation units [12••, 38, 39]. Current advances in genomics indicate that in the long term, genomic monitoring will be more limited by time and funding constrains in the assessment of demographic parameters and by conceptual and analytical weaknesses of biostatistical and bioinformatic tools, than by genotyping needs and the amount of sequence data available. Furthermore, advances in marker development and use, as well as in bioanalytical approaches and software (especially for non-model species, conifers in particular), will improve monitoring. However, in the 10–15 years between monitoring assessments, new markers and tools may be advanced, potentially complicating comparisons with data derived from previously used approaches. For this reason, DNA storage of all monitoring assessments is important as it allows a potential evaluation of earlier and current samples with state of the art markers and bioinformatic tools.
The application of genomic monitoring may bring forward a less blurry image of population status, but this image is still going to be far from fully clear. Most phenotypic traits involved in local adaptation are highly polygenic, and the importance of epistasis, transposable element activity [59] or epigenetics [64] has been suggested. Determining the contribution of epigenetic variation to phenotypic variability is still elusive; however, the future of genetic monitoring may have an epigenetic or epigenomic component. The distinction between phenotypic plasticity and adaptive genetic responses and the detection of signatures of selection in tree genomes are still not that straightforward [3, 62, 65••, 66]. However, genomic monitoring will generally improve indicator and verifier estimation, and a better determination of the critical levels between temporal assessments, as well as a better evaluation of the genetic and demographic thresholds that lead to maladaptation [18•, 27]. Furthermore, the advent of metagenomics approaches can boost research into the genetic and co-evolutionary interactions across forest communities (insect metagenomics, soil metagenomics, etc.). Metagenomics can be an essential component of genetic monitoring, although an extension to this level will make the monitoring exercise even more complex, demanding and challenging in terms of human power and cost.
Conclusions
Conservation of forest genetic resources is needed more than ever, given the cumulative effects of forest exploitation and destruction that is compounded by climate change. The need for the establishment of gene conservation units for forest tree species is high, as is their genetic monitoring. Genetic monitoring becomes more imperative as the speed of environmental change increases. Recent theoretical developments call for the application of the geneecological approach for forest genetic monitoring and the use of a limited number of highly informative indicators and their verifiers (demographic parameters and genetic diversity estimates).
The application of genomics can open new ways for a substantial enhancement of forest tree conservation activities. Highly precise estimates of genetic and demographic parameters and high-resolution characterization of adaptive genetic variation are feasible in natura. Genomic data will improve the processes for delineating areas for conservation due to the prospect for quantifying adaptive patterns. Functional genomic variation and the identification of genome parts that are responsible for local adaptation, and therefore important to conserve, can readily be evaluated. Forest genomic monitoring will advance the effectiveness of genetic monitoring to an entirely higher level. The meticulous evaluation of natural selection and demographic effects by genomic monitoring, the potential for discriminating these effects and unravelling the association of genetic diversity and environmental gradients, will increase manifold the precision of the estimation of adaptive evolutionary potential. Advances in forest tree genomics will enhance our ability to evaluate both genetic and environmental threats and their interaction. They will lead to integrated conservation genomics and genomic monitoring, as well as to better informed and comprehensive forest policy and management, and towards a better management and protection of forest genetic resources.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
van Andel T, Zangger E, Demitrack A. Land use and soil erosion in prehistoric and historical Greece. J Field Archaeol. 1990;17:379–96.
UNEP. Biodiversity 2015. http://www.unep.org/dgef/Biodiversity, Accessed 20 Sept. 2015.
Aravanopoulos FA. Genomics of trees. In: Ramawat KG, Merillon JM, Ahuja MR, editors. Tree Biotechnology. Boca Raton. CRC Press 2014. pp. 514-557.
FAO. The State of the World’s Forest Genetic Resources. Commission on Genetic Resources for Food and Agriculture, Food and Agriculture Organization of the United Nations, Rome; 2014.
Loo J, Souvannavong O, Dawson IK. Seeing the trees as well as the forest: the importance of managing forest genetic resources. For Ecol Manag. 2014;333:1–8.
CBD—Convention on Biological Diversity. United Nations Treaty Series 1993; 1760: 143-382.
Graudal L, Aravanopoulos FA, Bennadji Z, Changtragoon S, Fady B, Kjaer ED, et al. Global to local genetic diversity indicators of evolutionary potential in tree species within and outside forests. For Ecol Manag. 2014;333:35–51. A comprehensive account and a proposal for the use of genetic diversity indicators (state, pressure, benefit and response indicators) for forests, agroforestry, plantations and farms.
McKinnel FH. Status and trends in indicators of forest genetic diversity. FGR 38, Food and Agriculture Organization of the United Nations, Rome; 2002.
Laikre L. Genetic diversity is overlooked in international conservation policy implementation. Conserv Genet. 2010;11:349–54.
Aravanopoulos FA. Genetic monitoring in natural perennial plant populations. Botany. 2011;89:75–81. A theoretical paper that attempted to formalize the forest genetic monitoring concept and approach, including its scientific basis and universal application, the system of criteria, indicators and verifiers and the use of a temporal approach.
Konnert M, Maurer W, Degen B, Katzel R. Genetic monitoring in forests—early warning and controlling system for ecosystemic changes. iForest. 2011;4:77–81. This paper describes the application of a forest genetic monitoring system, discusses implications and raises issues for discussion on future genetic monitoring applications.
Aravanopoulos, F.A., Tollefsrud, M.M., Graudal, L., Koskela, J., Kätzel, R., Soto, A., et al. . 2015. Development of genetic monitoring methods for genetic conservation units of forest trees in Europe. European Forest Genetic Resources Programme (EUFORGEN), Bioversity International, Rome, Italy, 62 pp. This report pertains to the EUFORGEN Genetic Monitoring Working Group recommended comprehensive methodology for the genetic monitoring of genetic conservation units of forest trees in Europe, including scientific, technical and financial considerations for on-site application.
IPCC Intergovernmental Panel on Climate Change. Climate Change 2013: The Physical Science Basis. In: Stocker TF, Qin D, Plattner GK, Tignor M, Allen SK, Boschung, Nauels A, Xia Y, Bex V, Midgley PM (editors). Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge, Cambridge University Press; 2013.
Saha P. Modern climatology. New Delhi: Allied Publ; 2012.
Pertoldi C, Bijlsma R, Loeschcke V. Conservation genetics in a globally changing environment: present problems, paradoxes and future challenges. Biodivers Cons. 2007;16:4147–63.
Caballero A. Developments in the prediction of effective population size. Heredity. 1994;73:657–79.
Courchamp F, Berec J, Gascoigne J. Allee effects in ecology and conservation. Oxford: Oxford University Press; 2008.
Fady B, Cottrell J, Ackzell L, Alia R, Muys B, Prada A, et al. Forests and global change: what can genetics contribute to the major forest management and policy challenges of the twenty-first century? Reg Environ Change. 2015. doi:10.1007/s10113-015-0843-9. A broad review on the main challenges facing the world’s forests and a detailed account on how recent progress in genetics may be used to implement forest policy and managers and promote forest resilience to climate change.
Frankham R. Genetics and extinction. Biol Cons. 2005;126:131–40.
Falconer DS, MackayTFC. Introduction to quantitative genetics. 4th ed. Edinburgh, Longman, 1996.
Slatkin M. Gene flow and the geographic structure of natural-populations. Science. 1987;236:787–92.
Kremer A, Ronce O, Robledo-Arnuncio JJ, Guillaume F, Bohrer G, Nathan R, et al. Long-distance gene flow and adaptation of forest trees to rapid climate change. Ecology Letters. 2012;15:378–92.
Lande R, Shannon S. The role of genetic variation in adaptation and population persistence in changing environment. Evolution. 1996;50:434–7.
Namkoong G, Boyle T, Gregorious HR, Joly H, Savolainen O, Ratman W, Young A. Testing criteria and indicators for assessing the sustainability of forest management: genetic criteria and indicators. Centre for International Forestry Research, Working paper No. 10, Bogor; 1996.
Rajora OP, Mosseler A. Challenges and opportunities for conservation of forest genetic resources. Euphytica. 2001;118:197–212.
Kramer AT, Havens K. Plant conservation genetics in a changing world. Trends Plant Sci. 2009;14:599–607.
Aravanopoulos FA, Ganopoulos I, Tsaftaris A. Population and conservation genomics in forest and fruit trees. Adv Bot Res. 2015;74:125–56.
Ryman N, Palm S, Andre C, Carvalho CR, Dahlgren TG, Jorde PE, et al. Power for detecting genetic divergence: differences between statistical methods and marker loci. Mol Ecol. 2006;15:231–45.
Lande R. Genetics and demography in biological conservation. Science. 1988;241:1455–60.
Stebbins GL, Hartl DL. Comparative evolution: latent potentials for anagenetic advance. Proc Natl Acad Sci USA. 1988;85:5141–5.
Kimura M. The neutral theory of molecular evolution. Cambridge: Cambridge Univ. Press; 1983.
McKay JK, Latta RG. Adaptive population divergence: markers. QTL and traits Trends Ecol Evol. 2002;17:285–91.
Mittell EA, Nakagawa S, Hadfield JD. Are molecular markers useful predictors of adaptive potential? Ecol Lett. 2015. doi:10.1111/ele.12454.
Leinonen T, O’Hara RB, Cano JM, Merila J. Comparative studies of quantitative trait and neutral marker divergence: a meta-analysis. J Evol Biol. 2008;21:1–17.
Reed DH, Frankham R. Correlation between fitness and genetic diversity. Cons Biol. 2003;17:230–7.
Lynch M, Lande R. The critical effective size for a genetically secure population. Anim Conserv. 1998;1:70–2.
Kristensen TN, Sorensen AC. Inbreeding—lessons from animal breeding, evolutionary biology and conservation genetics. Animal Sci. 2005;80:121–33.
Funk WC, McKay JK, Hohenlohe PA, Allendorf FW. Harnessing genomics for delineating conservation units. Trends Ecol Evol. 2012;27:489.
Ouborg NJ, Angeloni F, Vergeer P. An essay on the necessity and feasibility of conservation genomics. Cons Genet. 2010;11:643–53.
Hoban SM, Hauffe HC, Perez-Espona S, Arntzen JW, Bertorelle G, Bryja J, et al. Bringing genetic diversity to the forefront of conservation policy and management. Cons Genet Res. 2013;5:593–8.
McMahon BJ, Teeling EC, Hoglund J. How and why should we implement genomics into conservation? Evol Appl. 2014;7:999–1007.
Schueler S, Falk W, Koskela J, Lefevre F, Bozzano M, Hubert J, et al. Vulnerability of dynamic genetic conservation units of forest trees in Europe to climate change. Global Change Biol. 2014;20:1498–511.
Sork VL, Aitken SN, Dyer RJ, Eckert AJ, Legendre P, Neale DB. Putting the landscape into the genomics of trees: approaches for understanding local adaptation and population responses to changing climate. Tree Genet Genomes. 2013;9:901–11.
Narum SR, Buerkle CA, Davey JW, Miller MR, Hohenlohe PA. Genotyping-by-sequencing in ecological and conservation genomics. Mol Ecol. 2013;22:2841–7.
Neale DB, Kreme A. Forest tree genomics, growing resources and applications. Nature Rev Genet. 2011;12:111–22. A review on the developments of forest tree genomics and their potential applications in the study of tree adaptation to the environment.
Savolainen O, Lascoux M, Merila J. Ecological genomics of local adaptation. Nat Rev Genet. 2013;14:807–20. An excellent review on the genetic basis of local adaptation and the development of research strategies that combine genomics and field experiments for its study.
Plomion C, Bastien C, Bogeat-Triboulot M-B, Bouffier L, Déjardin A, Duplessis S, et al. Forest tree genomics: 10 achievements from the past 10 years and future prospects. Annals For Sci. 2015;73:77–103.
Allendorf FW, Ryman N. Genetic management of hatchery stocks. In: Ryman N, Utter FM, editors. Population genetics and fishery management. Seattle: Univ. Washington Press; 1987. p. 141–59.
Schwartz MK, Luikart G, Waples RS. Genetic monitoring as a promising tool for conservation and management. Trends Ecol Evol. 2006;22:25–33.
Kuparinen A, Merila J. Detecting and managing fisheries-induced evolution. Trends Ecol Evol. 2007;22:652–9.
Allendorf FW, England PR, Luikart G, Ritchie PA, Ryman N. Genetic effects of harvest on wild animal populations. Trends Ecol Evol. 2008;23:327–37.
Laikre L, Larsson LC, Palme A, Charlier J, Josefsson M, Ryman N. Potentials for monitoring gene level biodiversity: using Sweden as an example. Biodivers Conserv. 2008;17:893–910.
Arandjelovic M, Head J, Kohl H, Boesch C, Robbins MM, Maisels F, et al. Effective non-invasive genetic monitoring of multiple wild western gorilla groups. Biol Cons. 2010;143:1780–91.
Geburek T, Milasowsky N, Frank C, Konrad H, Schadauer K. The Austrian forest biodiversity index: all in one. Ecol Indic. 2010;10:753–61.
Feld CK, da Silva PM, Sousa JP, et al. Indicators of biodiversity and ecosystem services: a synthesis across ecosystems and spatial scales. Oikos. 2009;118:1862–71.
Hampe A, Petit RJ. Conserving biodiversity under climate change: the rear edge matters. Mol Ecol. 2005;8:461–7.
Moritz C. Conservation units and translocations: strategies for conserving the evolutionary process. Hereditas. 1999;130:217–28.
Graudal L, Kjær E, Thomsen A, Larsen AB. Planning national programmes for conservation of forest genetic resources. Tech. Note No. 48. Humlebk, Danida Forest Seed Centre; 1997.
Grattapaglia D, Plomion C, Kirst M, Sederoff RR. Genomics of growth traits in forest trees. Curr Opinion Plant Biol. 2009;12:148–56.
Cullingham CI, Cooke JEK, Coltman DW. Cross-species outlier detection reveals different evolutionary pressures between sister species. New Phytologist. 2014;204:215–29.
Gonzalez-Martinez SC, Krutovsky KV, Neale DB. Forest tree population genomics and adaptive evolution. New Phytol. 2006;170:227–38.
Savolainen O, Pyhajarvi T. Genomic diversity in forest trees. Curr Opinion Plant Biol. 2007;10:162–7.
Antao T, Perez-Figueroa A, Luikart G. Early detection of population declines: high power of genetic monitoring using effective population size estimators. Evol Appl. 2011;4:144–54.
Avramidou EV, Ganopoulos IV, Doulis AG, Tsaftaris AS, Aravanopoulos FA. Beyond population genetics: natural epigenetic variation in wild cherry (Prunus avium). Tree Genet Genomes. 2015;11:95. doi:10.1007/s11295-015-0921-7.
Hansen MM, Olivieri I, Waller DM, Nielsen EE, the Genetic Monitoring Group, Allendorf FW, et al. Monitoring adaptive genetic responses to environmental change. Mol Ecol. 2012;21:1311–29. This insightful review discusses the use of genetic monitoring in the study of adaptive responses by both phenotypic and molecular genetics approaches. It proposes criteria for demonstrating adaptive responses by establishing clear links between genetic and environmental change.
Neale DB, Ingvarsson PK. Population, quantitative and comparative genomics of adaptation in forest trees. Curr Opinion Plant Biol. 2008;11:149–55.
Acknowledgments
Partial assistance from the European Union LIFE+ Project “LIFE for European Forest Genetic Monitoring System” is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Dr Aravanopoulos has no conflicts of interests to declare.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by the author.
Additional information
This article is part of the Topical Collection on Tree Genetics
Rights and permissions
About this article
Cite this article
Aravanopoulos, F.A.(. Conservation and Monitoring of Tree Genetic Resources in Temperate Forests. Curr Forestry Rep 2, 119–129 (2016). https://doi.org/10.1007/s40725-016-0038-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40725-016-0038-8