Abstract
Background and Objective
Pathogenic mutations in BRCA1/2 tumor suppressor genes increase the lifetime risk for developing breast and ovarian cancer. The aim of the present study was to evaluate the sensitivity and specificity of the Ion Torrent PGM™ for diagnostic mutation screening of BRCA1/2 genes.
Methods
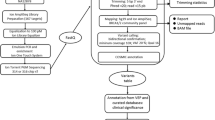
In the current study we included a cohort of 58 Bulgarian high-risk breast cancer patients to validate a next-generation sequencing approach for diagnostic mutation screening of the BRCA1/2 genes using the Ion Torrent Personal Genome Machine® (PGM™) platform. We have also optimized the workflow by comparing two different library preparation methods and using three software packages: NextGENE, Torrent Suite variantCaller, and Samtools/BCFtools to achieve detection of all variants with high specificity and sensitivity.
Results
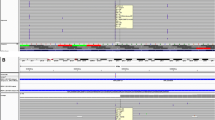
We have validated a quick and accurate diagnostic test, with an overall specificity of 95.9 % and sensitivity of up to 100 %, which can be the first method of choice, followed by confirmation of the identified variants by Sanger sequencing. Our results prove that the Ion AmpliSeq™ BRCA1/2 Community Panel used with the PGM™ platform, and coupled with our variant selection pipeline, is able to detect all sequence variants discovered by Sanger sequencing.
Conclusion
The application of the new test which outperforms the classical approach in turn-around time and price will have great impact in the clinical practice to identify the mutation carriers and guide the better personalized treatment of the patients, as well as to contribute for the improved prophylaxis in hereditary breast and ovarian cancer families.



Similar content being viewed by others
References
National Cancer Institute, NIH. BRCA1 and BRCA2: Cancer Risk and Genetic Testing [Internet]. 2014. Available from: http://www.cancer.gov/cancertopics/causes-prevention/genetics/brca-fact-sheet.
National Comprehensive Cancer Network. Genetic/Familial High-Risk Assessment: Breast and Ovarian (2013) [Internet]. Available from: http://www.nccn.org/professionals/physician_gls/f_guidelines.asp#breast_risk.
Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72:1117–30.
Chen S, Parmigiani G. Meta-analysis of BRCA1 and BRCA2 penetrance. J Clin Oncol. 2007;25:1329–33.
Kobayashi H, Ohno S, Sasaki Y, Matsuura M. Hereditary breast and ovarian cancer susceptibility genes (review). Oncol Rep. 2013;30:1019–29.
Karami F, Mehdipour P. A comprehensive focus on global spectrum of BRCA1 and BRCA2 mutations in breast cancer. Biomed Res Int. 2013;2013:928562.
National Human Genome Research Institute. Breast Cancer Information Core [Internet]. 2000. Available from: http://www.genome.gov/10001504.
De Leeneer K, Coene I, Poppe B, De Paepe A, Claes K. Rapid and sensitive detection of BRCA1/2 mutations in a diagnostic setting: comparison of two high-resolution melting platforms. Clin Chem. 2008;54:982–9.
Marsh DJ, Howell VM. The use of denaturing high performance liquid chromatography (DHPLC) for mutation scanning of hereditary cancer genes. Methods Mol Biol. 2010;653:133–45.
Mattocks CJ, Watkins G, Ward D, Janssens T, Bosgoed EA, van der Donk K, et al. Interlaboratory diagnostic validation of conformation-sensitive capillary electrophoresis for mutation scanning. Clin Chem. 2010;56:593–602.
Gargis AS, Kalman L, Berry MW, Bick DP, Dimmock DP, Hambuch T, et al. Assuring the quality of nextgeneration sequencing in clinical laboratory practice. Nat Biotechnol. 2012;30:1033–6.
Rehm HL, Bale SJ, Bayrak-Toydemir P, Berg JS, Brown KK, Deignan JL, et al. ACMG clinical laboratory standards for next-generation sequencing. Genet Med. 2013;15:733–47.
Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, et al. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol. 2012;30:434–9.
Costa JL, Sousa S, Justino A, Kay T, Fernandes S, Cirnes L, et al. Nonoptical massive parallel DNA sequencing of BRCA1 and BRCA2 genes in a diagnostic setting. Hum Mutat. 2013;34:629–35.
Bosdet IE, Docking TR, Butterfield YS, Mungall AJ, Zeng T, Coope RJ, et al. A clinically validated diagnostic second-generation sequencing assay for detection of hereditary BRCA1 and BRCA2 mutations. J Mol Diagn. 2013;15:796–809.
Tarabeux J, Zeitouni B, Moncoutier V, Tenreiro H, Abidallah K, Lair S, et al. Streamlined ion torrent PGMbased diagnostics: BRCA1 and BRCA2 genes as a model. Eur J Hum Genet. 2014;22:535–41.
Feliubadaló L, Lopez-Doriga A, Castellsagué E, del Valle J, Menéndez M, Tornero E, et al. Next-generation sequencing meets genetic diagnostics: development of a comprehensive workflow for the analysis of BRCA1 and BRCA2 genes. Eur J Hum Genet. 2013;21:864–70.
Weitzel J, Costa J, Mensenkamp A, Ekici A, Herzog J, Ligtenberg M, et al. BRCA1 and BRCA2 (BRCA) gene analyses on an economic platform: a global consortium to demonstrate the feasibility of a shared, dedicated workflow for non-optical next generation sequencing (NGS) with a custom BRCA AmpliSeq™ kit on the Ion Torrent PGMTM. Meet. Am. Soc. Hum. Genet. Boston; 2013.
Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol Clifton NJ. 2000;132:365–86.
Helmholtz Center Munich, Institute of Human Genetics. ExonPrimer [Internet]. Available from: http://ihg.gsf.de/ihg/ExonPrimer.html.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9.
Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013;Chapter 7:Unit 7.20.
Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7:e46688.
Milne I, Stephen G, Bayer M, Cock PJ, Pritchard L, Cardle L, et al. Using Tablet for visual exploration of second-generation sequencing data. Brief Bioinform. 2013;14:193–202.
Bragg LM, Stone G, Butler MK, Hugenholtz P, Tyson GW. Shining a light on dark sequencing: characterising errors in ion Torrent PGM data. PLoS Comput Biol. 2013;9:e1003031.
Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, et al. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics. 2012;13:341.
International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–96.
Meldrum C, Doyle MA, Tothill RW. Next-generation sequencing for cancer diagnostics: a practical perspective. Clin Biochem Rev. 2011;32:177–95.
Acknowledgments
DD, RD, IP, TG, AM, VM and RK declare no conflicts of interest.
This study was supported by Grant DUNK 01/2/2009 “National University Complex in Biomedical and Translational Research”, funded by the National Science Fund, Ministry of Education, Youth and Science, Bulgaria and Grant 48, funded by the Science Fund of the Medical University of Sofia, Bulgaria. We are grateful to all our patients and clinical teams of Prof. Svetlana Christova and Prof. Konstanta Timcheva.
Declaration of ethical conduct
This study and all Grants that provided funding for it have been approved by the Medical Ethics Committee of the Medical University of Sofia. All patients have been properly informed and have signed an informed consent form prior inclusion in the study.
Author contributions
Study concept—RK and AM; molecular genetic analysis—DD, RD and TG; sequence alignment and data analysis—IP and TG; clinical and molecular interpretations of results—DD, IP, AM and RK; manuscript preparation—DD and IP; final critical revision—AM, RK and VM; design and coordination of the study—DD and RK; administrative supervision and support—VM; the guarantee for the overall content—DD and RK.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dacheva, D., Dodova, R., Popov, I. et al. Validation of an NGS Approach for Diagnostic BRCA1/BRCA2 Mutation Testing. Mol Diagn Ther 19, 119–130 (2015). https://doi.org/10.1007/s40291-015-0136-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-015-0136-5