Abstract
Mitochondria are cellular powerhouses and central organelles to the regulation of many biological processes, including cell death and metabolism, with mitochondrial dysfunction being a hallmark of many different diseases. MicroRNAs (miRNAs) are a growing class of endogenous non-coding RNAs, which function as master regulators and fine-tuners of the genome, primarily via post-transcriptional mechanisms. miRNAs and RNA interference components have recently been demonstrated to be present in mitochondria from several species. However, miRNA transport mechanisms, biological targets, and function at the mitochondrial level are not well understood, and are only now beginning to emerge. In this review, we describe the biogenesis of miRNAs and present the major findings regarding miRNAs localized to mitochondria, their origin and their putative biological function(s).
Similar content being viewed by others
Introduction
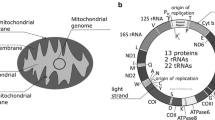
Mitochondria are pivotal organelles in cellular homeostasis, playing a central role in the regulation of multiple crucial cellular processes, including energy metabolism, cell viability and death, autophagy, and calcium trafficking, among others. Importantly, mitochondrial dysfunction or dysregulation is associated with multiple diseases, including cancer, neurodegenerative diseases, cardiomyopathies, metabolic syndrome, and obesity [1]. Human mitochondria possess a compact 16,569 bp circular genome, which is devoid of introns and codes for 13 protein subunits of the electron transport chain, and for multiple non-coding RNAs. Importantly, replication and transcription of mitochondrial DNA (mtDNA) are initiated from the small non-coding region, known as the D loop. These processes appear to be regulated primarily by proteins encoded in the nuclear genome, following post-translational import into mitochondria [1, 2••]. In turn, mtDNA transcription, translation, and transcript processing are regulated by multiple types of non-coding RNAs, encoded either in the mitochondrial genome or the nuclear genome following their uptake into mitochondria [3]. Interestingly, mitochondrial RNAs are transcribed from both strands, as long polycistronic precursor transcripts, and subsequently undergo processing, leading to the release of both non-coding and coding RNAs, including tRNAs, rRNAs, and mRNAs [2••].
MicroRNAs (miRNAs) have emerged as a class of non-coding RNAs processed from endogenous transcripts, functioning as master regulators and fine-tuners of the genome, via post-transcriptional mechanisms [4•, 5, 6, 7••]. miRNAs are capable of modulating the expression of multiple target genes and signaling pathways involved in the regulation of key cellular processes [7••], including cell growth and proliferation [8, 9], differentiation [10, 11], and apoptosis [8, 9, 12], as well as mitochondrial function (reviewed in [3, 13–16] ). Consequently, deregulation of a given miRNA can lead to the malfunction of pivotal cellular mechanisms, thus contributing to disease onset and/or progression. Therefore, it is not surprising that miRNA modulation has been increasingly demonstrated as a relevant therapeutic strategy in human disease [4, 8, 9, 12, 17].
miRNA Biogenesis and Mechanism(s) of Action
The discovery of miRNAs over two decades ago opened the door to a novel field of science. It has significantly changed our understanding of gene expression, providing an additional level of complexity to an already highly intricate process. miRNAs were discovered in C. elegans, where the lin-4 gene, which regulates the timing of larval development, was found to express two small RNA transcripts, 22- and 61-nucleotides (nt) long. Interestingly, these small RNAs displayed complementarity to the 3′UTR of lin-14. The negative modulation of lin-14 was shown to result from lin-4-mediated post-transcriptional regulation, which decreased lin-14 protein, while maintaining mRNA expression [18, 19]. This landmark finding was unrecognized for several years, until let-7 miRNA was discovered and found to display complementarity to the 3′UTR of several genes [20].
Since then, our understanding of miRNA biogenesis and mechanisms of action, as well as their influence on normal and disease processes has increased exponentially. This has led to the identification of ~2,600 mature miRNAs in humans, expressed from ~1,900 precursors, and many more miRNAs in other organisms including, mammals, worms, fish, and plants (www.miRbase.org). Interestingly, miRNAs were initially predicted to post-transcriptionally regulate over 30 % of human genes [21], which is now recognized as a gross underestimate. It has recently been suggested that miRNAs may, in fact, regulate >60 % of all human genes [22].
miRNAs are single stranded non-coding RNAs (ssRNAs) ~22 nt in length, whose main function is to post-transcriptionally silence gene expression [23], although in certain instances, miRNAs have also been reported to up-regulate translation [24]. miRNAs undergo several processing steps to generate a mature and biologically active miRNA molecule. The majority of miRNAs arise from a much-studied canonical miRNA biogenesis pathway (Fig. 1). However, some miRNAs appear to be produced by alternate non-canonical miRNA biogenesis pathways. These include debranched introns which mimic the structural features of pre-miRNAs, and enter the miRNA-processing pathway without Drosha-mediated cleavage (mirtrons) [25], in addition to splicing-independent mirtron-like miRNAs that do not require standard miRNA components for biogenesis (simtrons) [26].
Canonical miRNA transcription is primarily mediated by RNA polymerase II, producing 5′-capped, 3′-polyadenylated primary miRNAs (pri-miRNAs), containing hairpin stem-loop structure(s), with mature miRNAs lying on one arm of these stem-loops. Following transcription, the pri-miRNA is processed by nuclear RNase III endonuclease Drosha, resulting in the production of a double-stranded hairpin structure, termed precursor miRNA (pre-miRNA). In turn, pre-miRNAs are actively exported into the cytoplasm, via the Exportin-5 complex, where they are subsequently processed by the RNase III endonuclease Dicer, which cleaves near the terminal loop of the pre-miRNA hairpin and forms the miRNA duplexes. These miRNA duplexes are then incorporated into the miRNA-induced silencing complex (miRISC), followed by duplex unwinding and strand selection, in which the mature antisense miRNA (guide strand) is transferred to Argonaute (AGO) proteins within the miRISC. In general, the passenger strand is degraded rapidly. Following complex assembly, mature miRNA guides miRISC to target mRNAs, leading to their post-transcriptional silencing, which may proceed via translational repression and/or mRNA degradation. Under conditions of near or perfect miRNA complementarity, the target mRNA may also be endonucleolytic cleaved. In addition to their cytoplasmic localization, mature miRNAs are also somewhat unexpectedly present in sub-cellular compartments, including mitochondria and the nucleus. The mechanisms of miRNA trafficking to and from mitochondria are primarily unknown. However, there is some evidence that PNPASE, a 3′/5′ exoribonuclease and poly-A polymerase, located in the mitochondrial intermembrane space, or other yet unidentified proteins, is involved in this process. No doubt, PNPASE has been shown to play a pivotal role in the import of small RNA components into the matrix. In turn, the mechanisms underlying miRNA nuclear import are also being explored. Specifically, importin 8 physically associates with Ago2 and appears to regulate the transport of mature miRNAs into the cell nucleus. In addition to mature miRNAs, pre-miRNAs are also present in mitochondria, suggesting that mitochondria may provide a platform allowing the assembly of signaling complexes involved in the control of transcriptional repression
In the canonical biogenesis pathway, miRNAs are transcribed by RNA polymerase II, giving rise to primary miRNAs (pri-miRNAs), which exhibit varying lengths from hundreds to thousands of nucleotides and are 5′ 7-methyl-guanosine capped and 3′ polyadenylated, similar to mRNAs [27]. Nevertheless, a small number of miRNAs are found interspersed among repetitive elements transcribed by RNA polymerase III [28]. Pri-miRNA transcripts often originate from intergenic regions, where the miRNA can reside within an exon or intron of a non-coding transcript. However, many mammalian miRNAs reside within introns of protein-coding genes and display a similar transcription pattern to that of the protein-coding gene [29, 30]. In addition, pri-miRNAs structures typically contain a stem of ~33 base pairs, a terminal loop (forming a hairpin structure), and flanking ssRNA regions [31].
Following transcription, pri-miRNAs undergo enzymatic cleavage at the stem of the hairpin structure, releasing a 60–70 bp RNA product termed precursor miRNA (pre-miRNA). It is within one of the arms of this hairpin that the mature miRNA sequence is located. Processing of the pri-miRNA takes place within the nucleus, by enzymatic cleavage mediated by the RNAse III endonuclease Drosha, and its cofactor, the double-stranded-RNA-(dsRNA)-binding protein DiGeorge syndrome critical region gene 8 (DGCR8). Together they form a large (~650 kDa in humans) protein complex, termed microprocessor [32]. DGCR8 binds to the stem of the pri-miRNA and functions as a molecular anchor, aiding Drosha, which then cleaves the pri-miRNA at ~11 bp from the junction of the stem to the ssRNA. This process generates a pre-miRNA [31] intermediate containing a 5′ phosphate and a 2-nt 3′ overhang [32, 33]. Curiously, Drosha is also responsible for negatively regulating its cofactor DGCR8, by cleaving RNA hairpins within one of the exons of DGCR8 mRNA. This regulation illustrates the effect of exonic location of a miRNA hairpin, whereby Drosha-mediated endonucleolytic processing can destabilize the transcript, thus reducing protein synthesis. In addition, this was also suggested as a possible mechanism for mRNA downregulation, in a microprocessor-dependent and miRNA-independent manner [34].
Once the pre-miRNA has been “cropped” from the pri-miRNA, it is ready to be exported from the nucleus into the cytoplasm for further processing. Nuclear export is mediated primarily by exportin-5 (Exp5), a member of the nuclear receptor family, following the recognition of the 2-nt 3′ overhang generated by Drosha processing. Exp-5 binds its nuclear cargo pre-miRNAs in a Ran guanosine triphosphate (Ran-GTP)-dependent manner and releases pre-miRNAs into the cytoplasm, following hydrolysis of GTP [35–37]. Upon release into the cytosol, the pre-miRNAs are substrates for the RNase III endonuclease Dicer, which acts at the opposite end from Drosha near the terminal loop of the pre-miRNA hairpin [38]. The resulting miRNA duplexes are then transferred to argonaute (AGO) proteins and form the RNA-induced silencing complex (RISC), in association with dsRNA-binding proteins, TAR (HIV trans-activator RNA) RNA-binding protein (TRBP), and possibly the protein kinase R-activating protein (PACT) [5, 39, 40]. Subsequently, one strand of the RNA duplex remains bound to AGO as the mature miRNA (guide strand), while the other strand (passenger strand) is usually removed and degraded [23]. Which strand is retained depends on the relative thermodynamic stability of the two ends of the duplex intermediate [41, 42]. In humans, there are four argonaute proteins, AGO1-4, which can bind miRNAs, allowing them to exert post-transcriptional gene regulation. AGO2, or slicer as it is termed, is the catalytic component of RISC, and the only known human AGO with the capacity to cleave mRNAs [43].
Mature miRNAs loaded onto miRISC guide the complex to target mRNAs, leading to their post-transcriptional silencing. A key feature of target recognition in animals usually involves perfect, or near perfect, base-pairing between the miRNA nucleotides at positions 2–7 on the 5′ end, referred to as the “seed” sequence, and the 3′-untranslated region target sites of mRNA transcripts [6, 7••, 44, 45]. Since the requirement for miRNA targeting is complementarity to a small number of nucleotides, it is not surprising that each miRNA is predicted to interact with hundreds of target genes. Remarkably, a similar number of genes can, in fact, be regulated by a single miRNA [22, 46]. In mammals, miRNA target recognition and binding occur primarily via incomplete complementarity between the miRNA and target mRNA leading to target gene silencing, which may proceed via translational repression and/or mRNA degradation. Degradation of target mRNA due to miRNA action does not lead to AGO cleavage, but rather to target mRNA deadenylation, decapping, and exonucleolytic degradation [4•, 44, 45]. This process involves AGO, GW182, and the cellular decapping and deadenylation machinery, CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes, and is thought to occur in so-called processing bodies (P-bodies). These intracellular foci are discrete sites, where translationally inactive mRNAs are often stored and may undergo decay. miRNAs, their mRNA targets, as well as AGO proteins have all been identified in P-bodies [44, 45, 47]. Endonucleolytic cleavage can occur in cases of nearly perfect complementarity between miRNA and its target mRNA [6, 7••, 48].
Recently, a number of reports have confirmed the somewhat unexpected finding that mature miRNAs also exist in other subcellular organelles and compartments, including the nucleus [49•, 50], nucleolus [51, 52], and mitochondria (Table 1) [2••, 53•, 54, 55•, 56, 57]. These findings have significantly expanded our knowledge base and underscore the potential relevance of miRNAs in health and disease.
Mitochondria-Localized miRNAs and Their Potential Biological Targets and Function
Mitochondria are eukaryotic organelles that harbor their own genome in the form of mitochondrial DNA (mtDNA). Replication and transcription of mtDNA are regulated by multiple proteins, and non-coding RNAs that are encoded and expressed from both the mitochondrial and nuclear genomes. Up to ~1,500 proteins are found in mitochondria, and yet, the mitochondrial genome only encodes 13 proteins [2••, 58]. Therefore, the vast majority of mitochondrial proteins are synthesized in the cytoplasm and subsequently imported into mitochondria [59]. In contrast, most non-coding RNAs that function in mitochondria are encoded within the mtDNA [60]. However, it is well known that mitochondria also traffic multiple RNA species. Interestingly, many relevant nuclear-encoded RNAs are imported into mitochondria, including tRNAs [61], the RNA components of RNAse MRP and RNAse P [62], and 5S rRNA [63], which is the most abundant RNA within mitochondria. In addition, mitochondria are also able to export mitochondrially encoded tRNAs into the cytoplasm [64]. Importantly, mitochondria contain multiple families of non-coding RNAs, including miRNA, snRNA, piRNA, srpRNA, snoRNA, and repeat associated small RNAs [2••, 57].
There appear to be a number of different mechanisms for protein import into mitochondria [58, 65]. In addition, determining the role of protein–protein interactions in the regulation of mitochondrial function [66] will contribute significantly to our understanding of the full impact of mitochondrial miRNA (mitomiRs) target gene regulation, and signaling pathways under miRNA modulation. Over the last 5 years, we have begun to decipher the mechanisms involved in mitochondrial import and export of RNA [67–69], but not specifically for miRNAs. For example, polynucleotide phosphorylase (PNPASE), a 3′-to-5′ exoribonuclease and poly-A polymerase located in the mitochondrial intermembrane space appears to play a key role in the import of small RNA components required for mtDNA replication and mtRNA processing. In addition, PNPASE regulates adenine nucleotide levels and mitochondrial homeostasis, at least partly by regulating RNA import to control the abundance of electron transport chain components [67, 68]. These studies would suggest that PNPASE, and/or additional yet unidentified transporters, may be involved in the mitochondrial trafficking of miRNAs. The recent observation that hundreds of mature miRNAs reside within the cell nucleus raises the possibility of transcriptional regulation of gene expression [49•]. It is now established that importin 8 physically associates with Ago2 and regulates the transport of mature miRNAs into the cell nucleus [70]. The potential functional role(s) for mature miRNAs in the nucleus has established a heretofore unrecognized regulation of gene expression [50, 71].
Mitochondria are key subcellular organelles for the maintenance of cellular homeostasis. Together with the extensive array of both mitochondrial- and nuclear-encoded proteins and RNAs, the mitochondrion may represent a critical site for miRNA-mediated gene regulation. The first evidence that RNAi components localize to mitochondria was provided almost a decade ago, by the demonstration that human Ago2 interacts with mitochondrial tRNAmet [64]. Recently, Ago2 and Ago3 were found to co-localize to mitochondria, extending these initial observations, and increasing the potential relevance of mitochondria as a novel sub-cellular niche for RNAi-mediated gene silencing [54, 56, 57]. Interestingly, less than a decade ago, small non-coding RNAs were identified from mitochondria and chloroplasts by sequence analysis [72]. Since then, miRNAs have been reported to be present in mitochondria isolated from rat [53•] and mouse liver [54], human skeletal primary muscle cells [55•], human cancer cell lines Hela [56, 57], HEK293 [57], and 143B cells [2••], and rat cardiac myocytes [73•]. In addition to mature miRNAs, it was perhaps not surprising that precursor miRNAs have also been identified in mitochondria [55•] (Fig. 1).
There is a growing interest in the identification of miRNAs in mitochondria and their putative roles in cellular homeostasis. In this regard, it was only recently that our laboratory identified mature nuclear-encoded miRNAs in highly purified and intact mitochondria from rat liver [53•]. It was most critical to rule out potential contamination of the isolated mitochondria with cytosolic components containing miRNAs, such as endoplasmic reticulum, possibly Golgi and free ribosomes. The isolated and purified rat liver mitochondria were processed to ensure that they were devoid of contaminating RNAs from the cytosol or other cellular organelles and compartments. Extensive purification steps coupled with the detection of mature miRNAs by microarray, and northern blot validation essentially confirmed that mature miRNAs were indeed present in highly purified mitochondria [53•].
We initially identified a unique profile of 15 mature miRNAs in isolated mitochondria. Functional analysis using Miranda and TargetScan algorithms, as well as Ingenuity Pathway Analysis, indicated that they were neither targeting mitochondrial genes nor nuclear RNAs encoding mitochondrial proteins. Rather, these heretofore unrecognized mitochondria-associated miRNAs were apparently involved in the regulation of crucial biological processes such as apoptosis, cell proliferation, and differentiation [53•]. In 2010, a signature profile comprising 20 miRNAs was identified in intact mitochondria isolated from mouse liver [54]. Most notably, their enrichment was independent of the total cellular miRNA abundance, suggesting that mitochondria possess a distinctive population of miRNAs involved in the modulation of mitochondria-specific, as well as general cellular functions. In fact, in silico evaluation of the putative targets of mitomiRs miR-705, miR-494, and miR-202-5p by multiple target-predicting algorithms returned hits related to mitochondria-specific functions, such as tryptophanyl-tRNA synthetase (WARS) and transcription factor A (Tfam).
The first studies reporting the detection of mitochondrial miRNAs in human cells occurred in 2011, in which mitochondria were isolated from human skeletal primary muscle cells [55•] and HeLa cancer cell line [56]. A total of 46 miRNAs were identified in mitochondria from human skeletal muscle and found to be potentially involved in myogenesis, inflammation, fibrosis, oncogenesis, and tumor suppression. Importantly, these authors showed for the first time that precursor miRNAs were also localized to mitochondria, since 2 pre-miRNAs (pre-miR-302a and pre-miR-let-7a) were detected, and validated by in situ hybridization with visualization by confocal microscopy.
In silico analysis of the putative targets of these miRNAs returned a large number of sequences mapping to the mitochondrial genome and many of them within mitochondrial regulatory genes. Interestingly, some putative miRNA target genes displayed multiple miRNA target sequences. From their data, the ND6 gene displayed 38 putative miRNA target sites, which was twice as many as CYTB and ND1. In addition, other genes such as ND4L, ND4, and COX1 also displayed multiple potential miRNA binding sites. Further, miRNA let-7b was predicted to target a number of potential mitochondrial gene products, including ATP6, ATP8, COX2, and ND5, whereas other let-7 family members also displayed putative targets. Collectively, the authors suggested that some miRNAs detected in mitochondria, such as let-7 family miRNAs (let-7b, c, d, e, f, i), as well as miR-133a, could be involved in the regulation of mitochondrial gene expression by targeting mitochondrial mRNAs [55•].
In a separate but confirmatory study [56], 13 highly abundant nuclear-encoded miRNAs were identified in mitochondria isolated from human HeLa cancer cells. These miRNAs were detectable following a rigorous isolation procedure for mitochondria, including RNAse A treatment to remove contaminating miRNAs associated with the outer mitochondrial membrane. Further, in silico analysis of the targets of four of these mitochondrial miRNAs, miR-328, miR-494, miR-513, and miR-638 indicated their involvement in the regulation of mitochondrial homeostasis.
Recently, the human mitochondrial transcriptome from 143B osteosarcoma cells was analyzed by deep sequencing of RNA from isolated mitochondria treated with RNAse A to prevent contamination from non-mitochondrial RNAs. The results indicated that mitochondria contain a diverse population of small RNAs, comprising ~3 % of the whole-cell small RNA population, including miRNAs [2••]. Further, deep sequencing of purified mitochondrial RNA demonstrated a large number of reads aligning only to the nuclear genome and suggesting that these nuclear RNAs were imported into mitochondria. However, there was a strong enrichment in genes associated with protein translation indicating a potential contamination with ribosomes attached to the outer mitochondrial membrane, thereby raising concerns about the validity of the data. To address this problem, the RNA content of whole mitochondria was compared to the RNA content of mitoplasts (mitochondrial preparations stripped of their outer mitochondrial membrane). Matched sequencing of these RNAs showed a selective depletion of nuclear-encoded mRNAs and tRNAs in mitoplasts, indicating that the majority of nuclear-encoded RNAs found in mitochondria were indeed associated with the outer mitochondrial membrane. Importantly, the three miRNAs showing the greatest enrichment in whole mitochondria preparations, miR-146a, miR-103, and miR-16 also showed depletion in mitoplasts [2••].
Another recent study evaluated small RNA populations by deep sequencing RNA from mitochondria isolated from human cancer cells, HEK293, and HeLa cells, again demonstrating the presence of unique populations of small RNAs associated with mitochondria [57]. Interestingly, the authors characterized putative novel miRNAs from unannotated small RNA sequences. This study showed the association of 428 and 327 known, and 196 and 13 putative, novel miRNAs to mitochondria of HEK293 and HeLa, respectively. In silico analysis of potential targets showed that miRNA associated with mitochondria may regulate critical cellular processes, such as RNA turnover, apoptosis, cell cycle, and nucleotide metabolism, in which mitochondria play significant roles. Further, in silico analysis suggested that known miRNAs mapped to the mitochondrial genome, namely hsa-miR-4461, hsa-miR-4463, hsa-miR-4484, and hsa-miR-4485, which aligned to the genome at positions, respectively, corresponding to ND4L, ND5, L-ORF, and 16S rRNA mitochondrial genes. Furthermore, the authors also reported that 7 putative novel miRNAs aligned to non-coding regions, tRNA, 12S rRNA, and also to ATP6, ND2, HVRI, COI, CytB, and ND1 genes [57]. Importantly, miRNA targeting of mitochondrial transcripts was already experimentally validated for the nuclear-encoded miR-181c, which upon mitochondrial translocation was shown to regulate the expression of the mitochondria-encoded cytochrome c oxidase subunit 1 (mt-cox1) in rat cardiac myocytes [73•].
In addition to miRNAs located within mitochondria (mitoplasm), the mitochondrion itself may provide a platform allowing the assembly of signaling complexes involved in the control of transcriptional repression, expanding the relevance of miRNAs localized to this organelle. In this regard, miRNAs encoded in the nucleus have been shown to be associated with the mitochondrial outer membrane [2••, 57], together with RNAi components, Ago2, and Ago3 [54, 56, 57] (Fig. 1). In fact, miRNAs may be more significantly associated with the outer membrane than within the double-membrane bound organelle. It will require additional studies to elucidate the identification and dynamics of miRNAs, and their associated proteins, in the various submitochondrial compartments. Interestingly, mitochondria also interact with P-bodies, cytoplasmic bodies relevant in mRNA decay, storage, and RNAi, including miRNA-mediated cellular effects [74, 75]. In addition, dysregulation of mitochondrial function with carbonyl cyanide p-chlorophenylhydrazone was demonstrated to significantly decrease miRNA-mediated activity, possibly due to disruption of RISC assembly, associated with delocalization of Ago2 from P-bodies. The authors suggested that RNAi defects may be involved in pathologies associated with mitochondrial deficiencies [74].
Although there are many unanswered questions, it is now well established that miRNAs as well as their RISC complex proteins are both associated with and localized within mitochondria. The final destination of these miRNAs may be key to their function, including the regulation of mitochondria-encoded mRNA or nuclear-encoded mRNA/protein complexes. But other heretofore unrecognized roles may also exist. For example, miRNAs that associate with the outer membrane may function to regulate the mRNA/protein level at a distant subcellular site. The role of the mitochondrion clearly extends beyond its role in energy metabolism, inflammation, or apoptosis [3, 15]. The cross talk of mitochondria with the Golgi, the nuclear membrane, and P-bodies almost invariably underscores the importance of miRNAs in both normal and pathological conditions.
Mitochondria are extremely motile cellular organelles, whose dynamics carry significant biological impact, and this is particularly relevant to neurons [76, 77]. These are long-lived, post-mitotic, polarized cells, with relatively small cell body, harboring dendrites with multiple branches, and a thin axon that can be very long. Due to these particular morphological features, neurons face several challenges in maintaining energy homeostasis, thus requiring specialized mechanisms to efficiently distribute mitochondria to sub-cellular locations where energy is in high demand. These include synaptic terminals and axonal branches, which undergo dynamic remodeling during neuronal development and in response to synaptic activity, thereby altering the dynamics of mitochondrial trafficking. Therefore, the efficient regulation of mitochondrial trafficking and anchoring is essential to their recruitment and relocation to meet altered metabolic requirements, in addition to removing and replacing aged and damaged mitochondria [77].
Interestingly, there is growing evidence to suggest that mitochondria are also subject to intercellular transfer via exosomes and tunneling nanotubes, thereby impacting the regulation of target cells [78–80]. Collectively, this may significantly expand the relevance of mitochondrial miRNAs. These intracellular and intercellular dynamics may be biologically relevant, since mitochondria are important regulators of cell survival and cell death, and are also markedly involved in the regulation of signaling pathways elicited by perturbations in homeostasis [81]. Although requiring experimental validation, it is conceivable that following mitochondrial intra- or intercellular transference, miRNAs transported along with mitochondria, may lead to local enrichment in mitochondrial miRNAs, which upon organelle damage results in release of mitochondrial miRNAs and RNAi proteins. In turn, this could lead to a shift in local transcript targeting, which may ultimately translate into a putative biological effect. Importantly, changes impacting on organism homeostasis may alter whole tissue, as well as mitochondrial-associated miRNA expression. For example, streptozotocin treatment of mice, which induced type 1 diabetes and mitochondria dysfunction, altered the expression patterns of liver miRNAs and also of mitochondria-associated miRNAs, suggesting their potential involvement in mitochondrial dysfunction [54].
Conclusions
There is growing evidence to support the role of miRNAs in the regulation of mitochondrial function. In addition, it is increasingly evident that previously unexpected sub-cellular compartments, such as mitochondria or the nucleus, are extremely relevant sites of miRNA action. However, it is not entirely clear as to whether some mitochondria-localized miRNAs are encoded in the mitochondrial genome or only the nuclear genome and later imported into the mitochondria. Nevertheless and regardless of their nuclear or mitochondrial provenance, the biological and pathophysiological implications of mitomiRs are incompletely explored. Based on recent studies, it is conceivable that mitochondria-localized miRNAs may function as a relevant class of post-transcriptional regulators or fine-tuners of the mitochondrial genome. It is also not entirely known if mtDNA is a target, a source, or both, for miRNAs localizing to mitochondria. However, there is growing evidence to suggest that mitochondria may be both a source of miRNAs and a regulatory target for nuclear-encoded and mitochondria-localized miRNAs. In addition, it will also be interesting to determine if mitochondria might act as a storage and/or transport unit for miRNAs and proteins, including RNAi regulatory proteins, localized at, or enclosed within, this organelle. Proteins associated with mitochondria differ with cell type and tissue, depending on a myriad of factors influencing the local environment, including energy and metabolic requirements. It is becoming apparent that mitochondrial miRNA content may be similarly cell type-specific and regulated by intracellular context. Further knowledge of miRNA mitochondrial transport mechanisms will undoubtedly expand the relevance of miRNAs in the (de)regulation of cellular homeostasis. Importantly, additional studies are required to elucidate the full spectrum of mitochondria-localized/associated miRNAs, as well as to ascertain the extent of their biological implications and impact on human health and disease. These studies will undoubtedly expand the growing list of clinical targets for therapeutic intervention.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Nunnari J, Suomalainen A (2012) Mitochondria: in sickness and in health. Cell 148(6):1145–1159
•• Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AM, Haugen E, Bracken CP, Rackham O, Stamatoyannopoulos JA, Filipovska A, Mattick JS (2011) The human mitochondrial transcriptome. Cell 146 (4):645–658. This study provides a comprehensive map of the human mitochondrial transcriptome by near-exhaustive deep sequencing of long and short RNA fractions from purified mitochondria, identifying novel small RNAs, including miRNAs
Tomasetti M, Neuzil J, Dong L (2014) MicroRNAs as regulators of mitochondrial function: role in cancer suppression. Biochim Biophys Acta 1840(4):1441–1453
• Pereira DM, Rodrigues PM, Borralho PM, Rodrigues CMP (2013) Delivering the promise of miRNA cancer therapeutics. Drug Discov Today 18(5–6):282–289. Detailed review on miRNA in vivo delivery and therapeutic potential in human cancer
Graves P, Zeng Y (2012) Biogenesis of mammalian microRNAs: a global view. Genom Proteom Bioinform 10(5):239–245
Pasquinelli AE (2012) MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet 13(4):271–282
•• Huntzinger E, Izaurralde E (2011) Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 12(2):99–110. Detailed and outstanding review on miRNA gene silencing mechanisms
Borralho PM, Kren BT, Castro RE, da Silva IB, Steer CJ, Rodrigues CMP (2009) MicroRNA-143 reduces viability and increases sensitivity to 5-fluorouracil in HCT116 human colorectal cancer cells. FEBS J 276(22):6689–6700
Borralho PM, Simoes AE, Gomes SE, Lima RT, Carvalho T, Ferreira DM, Vasconcelos MH, Castro RE, Rodrigues CMP (2011) miR-143 overexpression impairs growth of human colon carcinoma xenografts in mice with induction of apoptosis and inhibition of proliferation. PLoS One 6(8):e23787
Aranha MM, Santos DM, Solá S, Steer CJ, Rodrigues CMP (2011) miR-34a regulates mouse neural stem cell differentiation. PLoS One 6(8):e21396
Aranha MM, Santos DM, Xavier JM, Low WC, Steer CJ, Solá S, Rodrigues CMP (2010) Apoptosis-associated microRNAs are modulated in mouse, rat and human neural differentiation. BMC Genomics 11:514
Castro RE, Ferreira DM, Afonso MB, Borralho PM, Machado MV, Cortez-Pinto H, Rodrigues CMP (2013) miR-34a/SIRT1/p53 is suppressed by ursodeoxycholic acid in the rat liver and activated by disease severity in human non-alcoholic fatty liver disease. J Hepatol 58(1):119–125
Li P, Jiao J, Gao G, Prabhakar BS (2011) Control of mitochondrial activity by miRNAs. J Cell Biochem 113(4):1104–1110
Bienertova-Vasku J, Sana J, Slaby O (2013) The role of microRNAs in mitochondria in cancer. Cancer Lett 336(1):1–7
Sripada L, Tomar D, Singh R (2012) Mitochondria: one of the destinations of miRNAs. Mitochondrion 12(6):593–599
Bandiera S, Matégot R, Girard M, Demongeot J, Henrion-Caude A (2013) MitomiRs delineating the intracellular localization of microRNAs at mitochondria. Free Radic Biol Med 64:12–19
Oberg AL, French AJ, Sarver AL, Subramanian S, Morlan BW, Riska SM, Borralho PM, Cunningham JM, Boardman LA, Wang L, Smyrk TC, Asmann Y, Steer CJ, Thibodeau SN (2011) miRNA expression in colon polyps provides evidence for a multihit model of colon cancer. PLoS One 6(6):e20465
Wightman B, Ha I, Ruvkun G (1993) Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75(5):855–862
Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75(5):843–854
Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403(6772):901–906
Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120(1):15–20
Friedman RC, Farh KK, Burge CB, Bartel DP (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19(1):92–105
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297
Vasudevan S, Tong Y, Steitz JA (2007) Switching from repression to activation: microRNAs can up-regulate translation. Science 318(5858):1931–1934
Ruby JG, Jan CH, Bartel DP (2007) Intronic microRNA precursors that bypass Drosha processing. Nature 448(7149):83–86
Havens MA, Reich AA, Duelli DM, Hastings ML (2012) Biogenesis of mammalian microRNAs by a non-canonical processing pathway. Nucleic Acids Res 40(10):4626–4640
Cai X, Hagedorn CH, Cullen BR (2004) Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10(12):1957–1966
Borchert GM, Lanier W, Davidson BL (2006) RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol 13(12):1097–1101
Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A (2004) Identification of mammalian microRNA host genes and transcription units. Genome Res 14(10A):1902–1910
Baskerville S, Bartel DP (2005) Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 11(3):241–247
Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN (2006) Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 125(5):887–901
Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R (2004) The Microprocessor complex mediates the genesis of microRNAs. Nature 432(7014):235–240
Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ (2004) Processing of primary microRNAs by the Microprocessor complex. Nature 432(7014):231–235
Han J, Pedersen JS, Kwon SC, Belair CD, Kim YK, Yeom KH, Yang WY, Haussler D, Blelloch R, Kim VN (2009) Posttranscriptional crossregulation between Drosha and DGCR8. Cell 136(1):75–84
Bohnsack MT, Czaplinski K, Gorlich D (2004) Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10(2):185–191
Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U (2004) Nuclear export of microRNA precursors. Science 303(5654):95–98
Yi R, Qin Y, Macara IG, Cullen BR (2003) Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17(24):3011–3016
Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409(6818):363–366
Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R (2005) TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436(7051):740–744
Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN (2006) The role of PACT in the RNA silencing pathway. EMBO J 25(3):522–532
Khvorova A, Reynolds A, Jayasena SD (2003) Functional siRNAs and miRNAs exhibit strand bias. Cell 115(2):209–216
Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD (2003) Asymmetry in the assembly of the RNAi enzyme complex. Cell 115(2):199–208
Peters L, Meister G (2007) Argonaute proteins: mediators of RNA silencing. Mol Cell 26(5):611–623
Liu X, Fortin K, Mourelatos Z (2008) MicroRNAs: biogenesis and molecular functions. Brain Pathol 18(1):113–121
Carthew RW, Sontheimer EJ (2009) Origins and mechanisms of miRNAs and siRNAs. Cell 136(4):642–655
Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res 36(Database issue):D154–158
Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E (2006) mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev 20(14):1885–1898
Kim VN, Han J, Siomi MC (2009) Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10(2):126–139
• Park CW, Zeng Y, Zhang X, Subramanian S, Steer, CJ (2010) Mature microRNAs identified in highly purified nuclei from HCT116 colon cancer cells. RNA Biol 7(5):606–614. The first report on the identification of numerous, and perhaps hundreds of mature microRNAs located in highly purified nuclei from a mammalian cell
Zisoulis DG, Kai ZS, Chang RK, Pasquinelli AE (2012) Autoregulation of microRNA biogenesis by let-7 and Argonaute. Nature 486(7404):541–544
Politz JC, Hogan EM, Pederson T (2009) MicroRNAs with a nucleolar location. RNA 15(9):1705–1715
Li ZF, Liang YM, Lau PN, Shen W, Wang DK, Cheung WT, Xue CJ, Poon LM, Lam YW (2013) Dynamic localisation of mature microRNAs in human nucleoli is influenced by exogenous genetic materials. PLoS One 8(8):e70869
• Kren BT, Wong PY, Sarver A, Zhang X, Zeng Y, Steer CJ (2009) MicroRNAs identified in highly purified liver-derived mitochondria may play a role in apoptosis. RNA Biol 6(1):65–72. The first report of the identification and profiling of miRNAs in intact and highly purified mitochondria, following RNAse A treatment
Bian Z, Li LM, Tang R, Hou DX, Chen X, Zhang CY, Zen K (2010) Identification of mouse liver mitochondria-associated miRNAs and their potential biological functions. Cell Res 20(9):1076–1078
• Barrey E, Saint-Auret G, Bonnamy B, Damas D, Boyer O, Gidrol X (2011) Pre-microRNA and mature microRNA in human mitochondria. PLoS One 6(5):e20220. The first report of precursor miRNAs detected in mitochondria, validated by in situ hybrizidation
Bandiera S, Ruberg S, Girard M, Cagnard N, Hanein S, Chretien D, Munnich A, Lyonnet S, Henrion-Caude A (2011) Nuclear outsourcing of RNA interference components to human mitochondria. PLoS One 6(6):e20746
Sripada L, Tomar D, Prajapati P, Singh R, Singh AK (2012) Systematic analysis of small RNAs associated with human mitochondria by deep sequencing: detailed analysis of mitochondrial associated miRNA. PLoS One 7(9):e44873
Herrmann JM, Longen S, Weckbecker D, Depuydt M (2012) Biogenesis of mitochondrial proteins. Adv Exp Med Biol 748:41–64
Neupert W, Herrmann JM (2007) Translocation of proteins into mitochondria. Annu Rev Biochem 76:723–749
Entelis NS, Kolesnikova OA, Martin RP, Tarassov IA (2001) RNA delivery into mitochondria. Adv Drug Deliv Rev 49(1–2):199–215
Alfonzo JD, Soll D (2009) Mitochondrial tRNA import—the challenge to understand has just begun. Biol Chem 390(8):717–722
Puranam RS, Attardi G (2001) The RNase P associated with HeLa cell mitochondria contains an essential RNA component identical in sequence to that of the nuclear RNase P. Mol Cell Biol 21(2):548–561
Entelis NS, Kolesnikova OA, Dogan S, Martin RP, Tarassov IA (2001) 5 S rRNA and tRNA import into human mitochondria. Comparison of in vitro requirements. J Biol Chem 276(49):45642–45653
Maniataki E, Mourelatos Z (2005) Human mitochondrial tRNAMet is exported to the cytoplasm and associates with the Argonaute 2 protein. RNA 11(6):849–852
Dudek J, Rehling P, van der Laan M (2012) Mitochondrial protein import: common principles and physiological networks. Biochim Biophys Acta 1833(2):274–285
Gu Z, Li J, Gao S, Gong M, Wang J, Xu H, Zhang C, Wang J (2011) InterMitoBase: an annotated database and analysis platform of protein–protein interactions for human mitochondria. BMC Genomics 12:335
Wang G, Shimada E, Koehler CM, Teitell MA (2012) PNPASE and RNA trafficking into mitochondria. Biochim Biophys Acta 1819(9–10):998–1007
Wang G, Chen HW, Oktay Y, Zhang J, Allen EL, Smith GM, Fan KC, Hong JS, French SW, McCaffery JM, Lightowlers RN, Morse HC 3rd, Koehler CM, Teitell MA (2010) PNPASE regulates RNA import into mitochondria. Cell 142(3):456–467
Wang G, Shimada E, Zhang J, Hong JS, Smith GM, Teitell MA, Koehler CM (2012) Correcting human mitochondrial mutations with targeted RNA import. Proc Natl Acad Sci USA 109(13):4840–4845
Wei Y, Li L, Wang D, Zhang CY, Zen K (2014) Importin 8 regulates the transport of mature microRNAs into the cell nucleus. J Biol Chem 289(15):10270–10275
Salmanidis M, Pillman K, Goodall G, Bracken C (2014) Direct transcriptional regulation by nuclear microRNAs. Int J Biochem Cell Biol. http://dx.doi.org/10.1016/j.biocel.2014.03.010. Accessed 05 May 2014
Lung B, Zemann A, Madej MJ, Schuelke M, Techritz S, Ruf S, Bock R, Huttenhofer A (2006) Identification of small non-coding RNAs from mitochondria and chloroplasts. Nucleic Acids Res 34(14):3842–3852
• Das S, Ferlito M, Kent OA, Fox-Talbot K, Wang R, Liu D, Raghavachari N, Yang Y, Wheelan SJ, Murphy E, Steenbergen C (2012) Nuclear miRNA regulates the mitochondrial genome in the heart. Circulation Res 110(12):1596–1603. The first report of nuclear-encoded miRNAs regulating the expression of mitochondrial genes. The authors show that miR-181c is encoded in the nucleus, assembled in the cytoplasm, and translocated into the mitochondria of cardiac myocytes, regulating the expression of mt-COX1
Huang L, Mollet S, Souquere S, Le Roy F, Ernoult-Lange M, Pierron G, Dautry F, Weil D (2011) Mitochondria associate with P-bodies and modulate microRNA-mediated RNA interference. J Biol Chem 286(27):24219–24230
Ernoult-Lange M, Bénard M, Kress M, Weil D (2012) P-bodies and mitochondria: which place in RNA interference? Biochimie 94(7):1572–1577
Du H, Guo L, Yan S, Sosunov AA, McKhann GM, Yan SS (2010) Early deficits in synaptic mitochondria in an Alzheimer’s disease mouse model. Proc Natl Acad Sci USA 107(43):18670–18675
Sheng ZH (2014) Mitochondrial trafficking and anchoring in neurons: new insight and implications. J Cell Biol 204(7):1087–1098
Ahmad T, Mukherjee S, Pattnaik B, Kumar M, Singh S, Rehman R, Tiwari BK, Jha KA, Barhanpurkar AP, Wani MR, Roy SS, Mabalirajan U, Ghosh B, Agrawal A (2014) Miro1 regulates intercellular mitochondrial transport & enhances mesenchymal stem cell rescue efficacy. EMBO J 33(9):994–1010
Spees JL, Olson SD, Whitney MJ, Prockop DJ (2006) Mitochondrial transfer between cells can rescue aerobic respiration. Proc Natl Acad Sci USA 103(5):1283–1288
Lou E, Fujisawa S, Morozov A, Barlas A, Romin Y, Dogan Y, Gholami S, Moreira AL, Manova-Todorova K, Moore MA (2012) Tunneling nanotubes provide a unique conduit for intercellular transfer of cellular contents in human malignant pleural mesothelioma. PLoS One 7(3):e33093
Galluzzi L, Kepp O, Kroemer G (2012) Mitochondria: master regulators of danger signalling. Nat Rev Mol Cell Biol 13(12):780–788
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Borralho, P.M., Rodrigues, C.M.P. & Steer, C.J. Mitochondrial MicroRNAs and Their Potential Role in Cell Function. Curr Pathobiol Rep 2, 123–132 (2014). https://doi.org/10.1007/s40139-014-0047-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40139-014-0047-x