Abstract
Purpose of Review
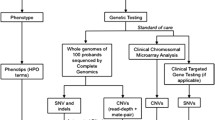
The purpose of this review was to compare existing strategies for evaluation of complex paediatric patients with newer techniques. Comparative genomic hybridization (CGH) array is the currently accepted first tier genetic test in the evaluation of a pediatric patient with complex physical and developmental anomalies. CGH provides an answer in only 15–20 % cases, and further genetic testing is required in the majority of cases. This has previously involved sequential single-gene tests, with low yield, significant costs and delay in diagnosis.
Summary of Recent Findings
New genetic techniques allowing massively parallel sequencing of multiple genes are becoming a part of medical practice as they provide a reduction in cost and time. Current medical practice supports the use of limited genomic testing—‘gene panels’ and ‘clinical exomes’, as a second tier approach after CGH array testing. These approaches have already been shown to improve the diagnostic yield providing an answer for an additional 25 % of patients. Ultimately, it is likely that whole genome sequencing as a single genomic test could replace CGH array and more restricted genomic tests, as research experience is translated into medical practice. Several factors need to be overcome to make this a reality to ensure equitable access to a reliable test with appropriate diagnostic interpretation.

Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: ∙ Of Importance
Lynch SA. What price a diagnosis? Dev Med Child Neurol. 2011;53:971.
Aoki Y, Niihori T, Inoue S-I, Matsubara Y. Recent advances in RASopathies. J Hum Genet. 2015;61:33–9.
Mikhail FM, Lose EJ, Robin NH, Descartes MD, Rutledge KD, Rutledge SL, et al. Clinically relevant single gene or intragenic deletions encompassing critical neurodevelopmental genes in patients with developmental delay, mental retardation, and/or autism spectrum disorders. Am J Med Genet. 2011;155:2386–96.
Finucane B, Challman TD, Martin CL, Ledbetter DH. Shift happens: family background influences clinical variability in genetic neurodevelopmental disorders. Genet Med. 2015;18:302–4.
Vissers L, Gilissen C, Veltman JA. Genetic studies in intellectual disability and related disorders. Nat Rev Genet. 2016;17:9–18.
Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–64.
Bejjani BA, Shaffer LG. Application of array-based comparative genomic hybridization to clinical diagnostics. J Mol Diagn. 2006;8:528–33.
Vissers LELM, de Vries BBA, Veltman JA. Genomic microarrays in mental retardation: from copy number variation to gene, from research to diagnosis. J Med Genet. 2010;47:289–97.
de Vries BBA, Pfundt R, Leisink M, Koolen DA, Vissers LELM, Janssen IM, et al. Diagnostic genome profiling in mental retardation. Am J Hum Genet. 2005;77:606–16.
Xiang B, Li A, Valentin D, Nowak NJ, Zhao H, Li P. Analytical and clinical validity of whole-genome oligonucleotide array comparative genomic hybridization for pediatric patients with mental retardation and developmental delay. Am J Med Genet. 2008;146A:1942–54.
Manning M, Hudgins L. Array-based technology and recommendations for utilization in medical genetics practice for detection of chromosomal abnormalities. Genet Med. 2010;12:742–5.
Kearney HM, South ST, Wolff DJ, Lamb A, Hamosh A, Rao KW. American College of Medical Genetics recommendations for the design and performance expectations for clinical genomic copy number microarrays intended for use in the postnatal setting for detection of constitutional abnormalities. Genet Med. 2011;13:676–9.
South ST, Lee C, Lamb AN, Higgins AW, Kearney HM. ACMG Standards and Guidelines for constitutional cytogenomic microarray analysis, including postnatal and prenatal applications: revision 2013. Genet Med. 2013;15:901–9.
Hanemaaijer NM, Sikkema-Raddatz B, van der Vries G, Dijkhuizen T, Hordijk R, van Essen AJ, et al. Practical guidelines for interpreting copy number gains detected by high-resolution array in routine diagnostics. Eur J Hum Genet. 2011;20:161–5.
Bruno D, Beddow R, Caramins M, Fagan K, Greville W, Harraway J, et al. Interpreting clinical microarray genomic data in 2012: what have we learnt and what challenges remain? Curr Top Genet. 2012;5:67–79.
Giorgio E, Ciolfi A, Biamino E, Caputo V, Di Gregorio E, Belligni EF, et al. Whole exome sequencing is necessary to clarify ID/DD cases with de novo copy number variants of uncertain significance: two proof-of-concept examples. Am J Med Genet. 2016;. doi:10.1002/ajmg.a.37649.
Gogarty B. Parents as Partners: a report and Guidelines on the investigations of children with developmental delay; by parents for professionals. Public health Genetics Unit, Cambridge Genetics Knowledge Park, 2006. Available at SSRN: http://ssrn.com/abstract=1796762.
∙ Muzzey D, Evans EA, Lieber C. Understanding the Basics of NGS: From Mechanism to Variant Calling. Curr Genet Med Rep. 2015;3:158–65. Review of technical aspects of Massively Parallel Sequencing techniques.
∙ Biesecker LG, Green RC. Diagnostic Clinical Genome and Exome Sequencing. N Engl J Med. 2014;370:2418–25. Review of technical aspects of Massively Parallel Sequencing techniques.
Nishiguchi K, Tearle R, Liu Y, Oh E, Katsanis N, Rivolta C. Whole genome sequencing in patients with retinitis pigmentosa reveals pathogenic DNA structuralchanges and NEK2 as a new disease gene. Proc Natl Acad Sci USA. 2013;10:16139–44.
Lupski JR, Gonzaga-Jauregui C, Yang Y, Bainbridge M, Jhangiani S, Buhay C, et al. Exome sequencing resolves apparent incidental findings and reveals further complexity of SH3TC2 variant alleles causing Charcot-Marie-Tooth neuropathy. Gen Med. 2013;5:57.
Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–6.
Mahdieh N, Rabbani B. An overview of mutation detection methods in genetic disorders. Iran J Pediatr. 2013;23:375–88.
Trump N, McTague A, Brittain H, Papandreou A, Meyer E, Ngoh A, et al. Improving diagnosis and broadening the phenotypes in early-onset seizure and severe developmental delay disorders through gene panel analysis. J Med Genet. 2016;53:310–7.
Shashi V, McConkie-Rosell A, Schoch K, Kasturi V, Rehder C, Jiang YH, et al. Practical considerations in the clinical application of whole-exome sequencing. Clin Genet. 2015;89:173–81.
∙ Wright CF, Fitzgerald TW, Jones WD, Clayton S, McRae JF, van Kogelenberg M, et al. Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. The Lancet. 2015;385:1305–14. Major study of exome sequencing in Development Disorders.
de Ligt J, Willemsen MH, van Bon BWM, Kleefstra T, Yntema HG, Kroes T, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367:1921–9.
Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med. 2013;369:1502–11.
Valencia CA, Husami A, Holle J, Johnson JA, Qian Y, Mathur A, et al. Clinical impact and cost-effectiveness of whole exome sequencing as a diagnostic tool: a pediatric center’s experience. Front Pediatr. 2015;3:121.
Monroe GR, Frederix GW, Savelberg SMC, de Vries TI, Duran KJ, van der Smagt JJ, et al. Effectiveness of whole-exome sequencing and costs of the traditional diagnostic trajectory in children with intellectual disability. Genetics in Medicine. 2016:1–8.
Stark Z, Tan TY, Chong B, Brett GR, Yap P, Walsh M, et al. A prospective evaluation of whole-exome sequencing as a first-tier molecular test in infants with suspected monogenic disorders. Genet Med. 2016;. doi:10.1038/gim.2016.1.
Tammimies K, Marshall CR, Walker S, Kaur G, Thiruvahindrapuram B, Lionel AC, et al. Molecular diagnostic yield of chromosomal microarray analysis and whole-exome sequencing in children with autism spectrum disorder. JAMA. 2015;314:895.
Soden SE, Saunders CJ, Willig LK, Farrow EG, Smith LD, Petrikin JE, et al. Effectiveness of exome and genome sequencing guided by acuity of illness for diagnosis of neurodevelopmental disorders. Sci Transl Med. 2014;6:265ra168.
van El CG, Cornel MC, Borry P, Hastings RJ, Fellmann F, Hodgson SV, et al. Whole-genome sequencing in health care. Eur J Hum Genet. 2013;21:S1–5.
Watson M. Incidental findings in clinical genomics: a clarification. Genetics in Medicine. 2013;15:664–6.
Hehir-Kwa JY, Claustres M, Hastings RJ, van Ravenswaaij-Arts C, Christenhusz G, Genuardi M, et al. Towards a European consensus for reporting incidental findings during clinical NGS testing. Eur J Hum Genet. 2015;23:1601–6.
Amendola LM, Jarvik GP, Leo MC, McLaughlin HM, Akkari Y, Amaral MD, et al. Performance of ACMG-AMP variant-interpretation guidelines among nine laboratories in the clinical sequencing exploratory research consortium. Am J Hum Genet. 2016;98:1067–76.
Jarvik GP, Browning BL. Consideration of cosegregation in the pathogenicity classification of genomic variants. Am J Hum Genet. 2016;98:1077–81.
Stavropoulos DJ, Merico D, Jobling R, Bowdin S, Monfared N, Thiruvahindrapuram B, et al. Whole-genome sequencing expands diagnostic utility and improves clinical management in paediatric medicine. Gen Med. 2016;1:15012.
Meienberg J, Bruggmann R, Oexle K, Matyas G. Clinical sequencing: is WGS the better WES? Hum Genet. 2016;135:359–62.
∙ Gilissen C, Hehir-Kwa JY, Thung DT, van de Vorst M, van Bon BWM, Willemsen MH, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–7. Major study of Whole Genome Sequencing in intellectual disability.
Mallawaarachchi AC, Hort Y, Cowley MJ, McCabe MJ, Minoche A, Dinger ME, et al. Whole-genome sequencing overcomes pseudogene homology to diagnose autosomal dominant polycystic kidney disease. Eur J Hum Genet. 2016;. doi:10.1038/ejhg.2016.48.
Ellingford J, Barton S, Bhaskar S, Williams SG, Sergouniotis PI, et al. Whole genome sequencing increases molecular diagnostic yield compared with current diagnostic testing for inherited retinal disease. Ophthalmology. 2016;123:1143–50.
Matthijs G, Souche E, Alders MEL, Corveleyn A, Eck S, Feenstra I, et al. Guidelines for diagnostic next-generation sequencing. Eur J Hum Genet. 2016;24:2–5.
ExAC Browser [Internet]. Broad Institute; [cited 2016 Jun 19]. http://exac.broadinstitute.org.
The Human Gene Mutation Database [Internet]. Institute of Medical Genetics in Cardiff; [cited 2016 Jun 19]. http://www.hgmd.cf.ac.uk/ac/index.php.
Dong C, Wei P, Jian X, Gibbs R, Boerwinkle E, Wang K, et al. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum Mol Genet. 2015;24:2125–37.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–23.
Maron BJ, Maron MS, Semsarian C. Genetics of hypertrophic cardiomyopathy after 20 years. JAC. 2012;60:705–15.
Beale S, Sanderson D, Sanniti A, Dundar Y, Boland A. A scoping study to explore the cost-effectiveness of next-generation sequencing compared with traditional genetic testing for the diagnosis of learning disabilities in children. Health Technol Assess. 2015;19:1–90.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
Amali Mallawaarachchi and Felicity Collins declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
This article is part of the Topical Collection on Genetics.
Rights and permissions
About this article
Cite this article
Mallawaarachchi, A., Collins, F. Testing the Complex Child: CGH Array, WES, Clinical Exome, WGS. Curr Pediatr Rep 4, 155–163 (2016). https://doi.org/10.1007/s40124-016-0111-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40124-016-0111-6