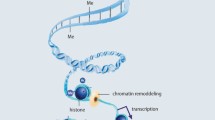
Summary
Not only genetics and environmental factors are responsible for the development of allergies. Epigenetic mechanisms – that are master regulators of gene transcription and thus of the accessibility and use of genome information – have been identified as a third power that is key in determining many features of allergy. So far, three epigenetic mechanisms have been identified in humans: histone modification, DNA methylation, and posttranscriptional modification by microRNA (miRNA) or small interfering RNA (siRNA). In recent years, it became clear that epigenetic regulation of gene function does not only play a role in the development of allergy but may also play a key role in the induction of tolerance against allergens, holding the promise to truly heal or prevent atopic diseases in the future.


Similar content being viewed by others
Abbreviations
- GSTM1:
-
Glutathione S-transferase M1
- GSTT1:
-
Glutathione S-transferase T1
- H:
-
Histone
- LPS:
-
Lipopolysaccharide
- MAPK:
-
Mitogen-activated protein kinase
- miRNA:
-
MicroRNA
- RNAi:
-
RNA interference
- siRNA:
-
Small interfering RNA
- SOTI:
-
Specific oral tolerance induction
- TLR:
-
Toll-like receptor
- Treg:
-
T regulatory cell
References
Bonnelykke K, Matheson MC, Pers TH, Granell R, Strachan DP, Alves AC et al. Meta-analysis of genome-wide association studies identifies ten loci influencing allergic sensitization. Nat Genet 2013;45:902–6
Weidinger S, Gieger C, Rodriguez E, Baurecht H, Mempel M, Klopp N et al. Genome-wide scan on total serum IgE levels identifies FCER1A as novel susceptibility locus. PLoS Genet 2008;4:e1000166
Ellinghaus D, Baurecht H, Esparza-Gordillo J, Rodriguez E, Matanovic A, Marenholz I, et al. High-density genotyping study identifies four new susceptibility loci for atopic dermatitis. Nat Genet 2013;45:808–12
Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 2007;448:470–3
Andiappan AK, Wang de Y, Anantharaman R, Parate PN, Suri BK, Low HQ et al. Genome-wide association study for atopy and allergic rhinitis in a Singapore Chinese population. PLoS One 2011;6:e19719.
Hong X, Hao K, Ladd-Acosta C, Hansen KD, Tsai HJ, Liu X et al. Genome-wide association study identifies peanut allergy-specific loci and evidence of epigenetic mediation in US children. Nat Commun 2015;6:6304
Kabesch M. Gene by environment interactions and the development of asthma and allergy. Toxicol Lett 2006;162:43–8
Riedler J, Braun-Fahrlander C, Eder W, Schreuer M, Waser M, Maisch S et al. Exposure to farming in early life and development of asthma and allergy: a cross-sectional survey. Lancet 2001;358:1129–33
Scholtens S, Postma DS, Moffatt MF, Panasevich S, Granell R, Henderson AJ et al. Novel childhood asthma genes interact with in utero and early-life tobacco smoke exposure. J Allergy Clin Immunol 2014;133:885–8
Kabesch M, Hoefler C, Carr D, Leupold W, Weiland SK, von Mutius E. Glutathione S transferase deficiency and passive smoking increase childhood asthma. Thorax 2004;59:569–73
Gilliland FD, Li YF, Dubeau L, Berhane K, Avol E, McConnell R et al. Effects of glutathione S-transferase M1, maternal smoking during pregnancy, and environmental tobacco smoke on asthma and wheezing in children. Am J Respir Crit Care Med 2002;166:457–63
Kormann MS, Depner M, Hartl D, Klopp N, Illig T, Adamski J et al. Toll-like receptor heterodimer variants protect from childhood asthma. J Allergy Clin Immunol 2008;122:86–92, 92 e1-8
Baldini M, Lohman IC, Halonen M, Erickson RP, Holt PG, Martinez FD. A Polymorphism* in the 5’ flanking region of the CD14 gene is associated with circulating soluble CD14 levels and with total serum immunoglobulin E. Am J Respir Cell Mol Biol 1999;20:976–83
Hinds DA, McMahon G, Kiefer AK, Do CB, Eriksson N, Evans DM et al. A genome-wide association meta-analysis of self-reported allergy identifies shared and allergy-specific susceptibility loci. Nat Genet 2013;45:907–11
Simpson A, John SL, Jury F, Niven R, Woodcock A, Ollier WE et al. Endotoxin exposure, CD14, and allergic disease: an interaction between genes and the environment. Am J Respir Crit Care Med 2006;174:386–92
Pandey RC, Michel S, Tesse R, Binia A, Schedel M, Liang L et al. Genetic variation in the Toll-like receptor signaling pathway is associated with childhood asthma. J Allergy Clin Immunol 2013;131:602–5
Tost J. Methods for the Genome-wide and Gene-specific Analysis of DNA Methylation Levels and Patterns. In: Horizon Scientific Press/ Caister Academic Press, Norwich, UK; 2008. p. pp. 63–104
Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res 2011;21:381–95
Bird A. DNA methylation patterns and epigenetic memory. Genes Dev 2002;16:6–21
Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005;120:15–20
Lal G, Zhang N, van der Touw W, Ding Y, Ju W, Bottinger EP et al. Epigenetic regulation of Foxp3 expression in regulatory T cells by DNA methylation. J Immunol 2009;182:259–73
Lee DU, Agarwal S, Rao A. Th2 lineage commitment and efficient IL-4 production involves extended demethylation of the IL-4 gene. Immunity 2002;16:649–60
Liang L, Willis-Owen SA, Laprise C, Wong KC, Davies GA, Hudson TJ, et al. An epigenome-wide association study of total serum immunoglobulin E concentration. Nature 2015;520:670–4
Rodríguez E, Baurecht H, Wahn AF, Kretschmer A, Hotze M, Zeilinger S et al. An integrated epigenetic and transcriptomic analysis reveals distinct tissue-specific patterns of DNA methylation associated with atopic dermatitis. J Invest Dermatol 2014;134:1873–83
Martino D, Joo JE, Sexton-Oates A, Dang T, Allen K, Saffery R, et al. Epigenome-wide association study reveals longitudinally stable DNA methylation differences in CD4+ T cells from children with IgE-mediated food allergy. Epigenetics 2014;9:998–1006
Syed A, Garcia MA, Lyu SC, Bucayu R, Kohli A, Ishida S, et al. Peanut oral immunotherapy results in increased antigen-induced regulatory T-cell function and hypomethylation of forkhead box protein 3 (FOXP3). J Allergy Clin Immunol 2014;133:500–10
DeWoskin VA, Million RP. The epigenetics pipeline. Nat Rev Drug Discov 2013;12:661–2
Author information
Authors and Affiliations
Corresponding author
Additional information
Conflict of interest
The author declares that there are no conflicts of interest.
Annotation
This review article is based on a presentation at the German Allergy Congress 2015.
Cite this as
Kabesch M. Epigenetic determinants of allergy and tolerance. Allergo J Int 2016;25:154–9 DOI: 10.1007/s40629-016-0122-4
Rights and permissions
About this article
Cite this article
Kabesch, M. Epigenetic determinants of allergy and tolerance. Allergo J 25, 32–37 (2016). https://doi.org/10.1007/s15007-016-1170-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s15007-016-1170-2