Abstract
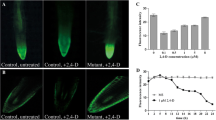
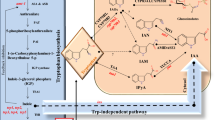
Auxins are a group of phytohormones that regulate several aspects of plant growth and development. Indole-3-acetic acid (IAA) is the predominant form of auxin in plants and several IAA biosynthetic pathways have been previously proposed but remain genetically uncharacterized. One of the proposed pathways is the indole-3-pyruvic acid (IPyA) pathway, which is inferred to regulate key developmental processes such as apical hook formation and shade avoidance. Recent molecular evidence suggests the existence of the pathway in higher plants but remains unverified due to the elusive nature of IPyA in vitro. Extending on these recent advances, this research was aimed at investigating aspects of IPyA-dependent auxin biology in Pisum sativum (pea) using reverse genetics, expression profiling, and analytical techniques. As a result the genes PsTAR2, PsTAR 5g Mt 80, and PsTAR 5g Mt 90, which are inferred to encode key enzymes in the IPyA pathway, were cloned. On expression analysis PsTAR2 was found to be slightly heightened in response to IPyA-inducing conditions (shade) while IAA levels remained unaltered contrary to previous reports. Moreover, the inferred homologs PsTAR 5g Mt 80 and PsTAR 5g Mt 90 appeared down-regulated in the same conditions suggesting functional divergence in the gene family. Thus, PsTAR2 was thought to be solely responsible for regulating IPyA-dependent auxin synthesis. Consequently, using a reverse genetic approach, called TILLING, the PsTAR2 gene was mutated in order to study the down-stream effects of IPyA deficiency. The procedure is currently underway and in the process of isolating two novel pstar2 (IPyA) mutant lines consisting of a missense mutation (pstar2 4280) and a highly desired knockout mutation (pstar2 918). On completion the novel mutants are anticipated to be indispensable to future IPyA-auxin investigations in higher plants. In light of the unstable nature of IPyA, a protocol has been formulated using UPLC for fractioning followed by MS/MS analysis. This technique appears to be very promising as a robust IPyA detection protocol in plant extracts.




Similar content being viewed by others
Abbreviations
- AA:
-
Acetic acid
- ACN:
-
Acetonitrile
- DNA:
-
Deoxyribonucleic acid
- cDNA:
-
Complementary DNA
- EMS:
-
Ethylmethane sulfonate
- GC:
-
Gas chromatography
References
Badenoch-Jones J, Summons RE, Rolfe BG, Letham DS (1984) Phytohormones, rhizobium mutants, and nodulation in legumes. IV. Auxin metabolites in pea root nodules. J Plant Growth Regul 3:23–39
Ballaré CL (1999) Keeping up with the neighbours: phytochrome sensing and other signalling mechanisms. Trends Plant Sci 4:97–102
Benjamins R, Scheres B (2008) Auxin: the looping star in plant development. Annu Rev Plant Biol 59:443–465
Bialek K, Michalczuk L, Cohen JD (1992) Auxin biosynthesis during seed germination in Phaseolus vulgaris. Plant Physiol 100:509–517
Cooney TP, Nonhebel HM (1989) The measurement and mass spectral identification of indole-3- pyruvate from tomato shoots. Biochem Biophys Res Commun 162:761–766
Cooney TP, Nonhebel HM (1991) Biosynthesis of indole-3-acetic acid in tomato shoots: Measurement, mass-spectral identification and incorporation of indole-3-acetic acid, d- and l-tryptophan, indole-3-pyruvate and tryptamine. Planta 184:368–376
Cummings IG, Foo E, Weller JL, Reid JB, Koutoulis A (2008) Blue and red photoselective shadecloths modify pea height through altered blue irradiance perceived by the cry1 photoreceptor. J Hortic Sci Biotechnol 83:663–667
Kanyuka K, Praekelt U, Franklin KA, Billingham OE, Hooley R, Whitelam GC, Halliday KJ (2003) Mutations in the huge Arabidopsis gene affect a range of hormone and light responses. Plant J 35:57–70
Koshiba T, Kamiya Y, Iino M (1995) Biosynthesis of indole-3-acetic acid from l-tryptophan in coleoptile tips of maize (Zea mays L.). Plant Cell Physiol 36:1503–1510
Kurepin LV, Emery RJN, Pharis RP, Reid DM (2007) Uncoupling light quality from light irradiance effects in Helianthus annuus shoots: putative roles for plant hormones in leaf and internode growth. J Exp Bot 58:2145–2157
Lehman A, Black R, Ecker JR (1996) HOOKLESS1 an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell 85:183–194
Morelli G, Ruberti I (2000) Shade avoidance responses. Driving auxin along lateral routes. Plant Physiol 122:621–626
Nonhebel HM, Cooney TP, Simpson R (1993) The route, control and compartmentation of auxin synthesis. Aust J Plant Physiol 20:527–539
Raz V, Ecker JR (1999) Regulation of differential growth in the apical hook of Arabidopsis. Development 126:3661–3668
Steindler C, Matteucci A, Sessa G, Weimar T, Ohgishi M, Aoyama T, Morelli G, Ruberti I (1999) Shade avoidance responses are mediated by the ATHB-2 HD-Zip protein, a negative regulator of gene expression. Development 126:4235–4245
Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, DoleZal K, Schlereth A, Jurgens G, Alonso JM (2008) TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133:177–191
Tam YY, Normanly J (1998) Determination of indole-3-pyruvic acid levels in Arabidopsis thaliana by gas chromatography-selected ion monitoring-mass spectrometry. J Chromatogr A 800:101–108
Tao Y, Ferrer JL et al (2008) Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133:164–176
Tivendale ND, Davies NW, Molesworth PP, Davidson SE, Smith JA, Lowe EK, Reid JB, Ross JJ (2010). Reassessing the role of N-hydroxytryptamine in auxin biosynthesis. Plant Physiol 154:1957–1965
Yamada M, Greenham K, Prigge MJ, Jensen PJ, Estelle M (2009) The TRANSPORT INHIBITOR RESPONSE2 gene is required for auxin synthesis and diverse aspects of plant development. Plant Physiol 151:168–179
Žádníková P, Petracek J et al (2010) Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development 137:607–617
Zhao Y (2010) Auxin biosynthesis and its role in plant development. Ann Rev Plant Biol 61:49–64
Acknowledgements
We the authors would like to thank Anna University and its associated labs for their help and support towards this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that no conflict of interest exists.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bala, R.K., Murugesan, R., Subramanian, S. et al. Auxin biosynthetic intermediate genes and their role in developmental growth and plasticity in higher plants. J. Plant Biochem. Biotechnol. 26, 321–329 (2017). https://doi.org/10.1007/s13562-016-0394-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13562-016-0394-2