Abstract
The determination of a robust reference gene has become increasingly important since RT-qPCR used as a prominent technique for quantification of transcript in connection with their molecular and biological mechanisms. Only a few studies on reference genes have been conducted using Antarctic ice algae. In this work, 10 candidate reference genes of Chlamydomonas sp. ICE-L were evaluated for their stabilities. The results showed that the best references genes differed across the experimental samples. Based on NormFinder Analysis, EF-1α was the most suitable reference gene under the diurnal cycle, high light, high salinity and UV-B irradiation conditions, and GAPDH was the most stable gene under different light intensities. For all tested samples H2B was the best gene and 18S was the least. Pair-wise variation analysis revealed that H2B and EF-1α were the best gene combination for diurnal cycle and high light conditions. For different light intensities and high salinity samples, the best combinations were GAPDH + ACT and L32 + H2B, respectively. For UV-B irradiated samples, a minimum of three genes (EF-1α, L32 and 18S) were necessary for accurate normalization. Selecting appropriate reference gene was very important to achieve an accurate and reliable normalization of genes’ expression. These results provided guidelines for reference genes selection under different experimental conditions and also established a foundation for more accurate and widespread use of RT-qPCR in Chlamydomonas sp. ICE-L.


Similar content being viewed by others
References
Andersen CL, Jensen JL, Orntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245–5250
Bail AL, Dittami SM, Franco PO, Rousvoal S, Cock MJ, Tonon T, Charrier B (2008) Normalisation genes for expression analyses in the brown alga model Ectocarpus siliculosus. BMC Mol Biol 9:75. doi:10.1186/1471-2199-9-75
Bustin SA (2002) Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol 29:23–39
Bustin SA, Benes V, Nolan T, Pfaffl MW (2005) Quantitative real-time RT-PCR-a perspective. J Mol Endocrinol 34:597–601
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139:5–17
Dheda K, Huggett JF, Bustin SA, Johnson MA, Rook G, Zumla A (2004) Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques 37:112–119
Gachon C, Mingam A, Charrier B (2004) Real-time PCR: what relevance to plant studies? J Exp Bot 55:1445–1454
Gonzalez-Verdejo CI, Die JV, Nadal S, Jimenez-Marin A, Moreno MT, Roman B (2008) Selection of housekeeping genes for normalization by real-time RTPCR: analysis of Or-MYB1 gene expression in Orobanche ramose development. Anal Biochem 379:176–181
Gutierrez L, Mauriat M, Pelloux J, Bellini C, Van Wuytswinkel O (2008) Towards a systematic validation of references in real-time RT-PCR. Plant Cell 20:1734–1735
Hong SY, Seo PJ, Yang MS, Xiang F, Park CM (2008) Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biol 8:112
Hu R, Fan C, Li H, Zhang Q, Fu Y (2009) Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol Biol 10:93. doi:10.1186/1471-2199–10-93
Huggett J, Dheda K, Bustin S, Zumla A (2005) Real-time RT-PCR normalisation; strategies and considerations. Genes Immun 6:279–284
Jain M, Nijhawan A, Tyagi AK, Khurana JP (2006) Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun 345:646–651
Jiang HB, Liu YH, Tang PA, Zhou AW, Wang JJ (2010) Validation of endogenous reference genes for insecticide-induced and developmental expression profiling of Liposcelis bostsrychophila (Psocoptera: Liposcelididae). Mol Biol Rep 37:1019–1029
Lee PD, Sladek R, Greenwood CMT, Hudson TJ (2002) Control genes and variability: absence of ubiquitous reference transcripts in diverse mammalian expression studies. Genome Res 12:292–297
Lin YL, Lai ZX (2010) Reference gene selection for qPCR analysis during somatic embryogenesis in longan tree. Plant Sci 178:359–365
Liu C, Huang X, Wang X, Zhang X, Li G (2006) Phylogenetic studies on two strains of Antarctic ice algae based on morphological and molecular characteristics. Phycologia 45:190–198
Liu S, Zhang P, Cong B, Liu C, Lin X, Shen J, Huang X (2010) Molecular cloning and expression analysis of a cytosolic Hsp70 gene from Antarctic ice algae Chlamydomonas species. ICE-L Extremophiles 14:329–337
Mallona I, Lischewski S, Weiss J, Hause B, Egea-Cortines M (2010) Validation of reference genes for quantitative real-time PCR during leaf and flower development in Petunia hybrida. BMC Plant Biol 10:4
Miao JL, Li GY, Hou XG (2002) Study on induced synthesis of anti-UV substances in the antarctic algae. High Technol Lett 6:179–183, Published in Chinese
Mock T, Valentin K (2004) Photosynthesis and cold acclimation: molecular evidence from a polar diatom. J Phycol 40:732–741
Nicot N, Hausman JF, Hoffmann L, Evers D (2005) Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot 56:2907–2914
ProvasoliL (1968) Media and prospects for the cultivation ofmarine algae. In: Watanabe A, Hattori R (eds) Culture and collections of algae. Proceedings of US–Japan Conference, Hakone, pp 63–95
Reid KE, Olsson N, Schlosser J, Peng F, Lund ST (2006) An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol 6:27
Suzuki T, Higgins PJ, Crawford DR (2000) Control selection for RNA quantitation. Biotechniques 29:332–337
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 18;3 (7): RESEARCH0034.
Walker NJ (2002) Tech. Sight. A technique whose time has come. Science 296(5567):557–559
Zhang P, Liu S, Cong B, Wu G, Liu C, Lin X, Shen J, Huang X (2011) A novel omega-3 fatty acid desaturase involved in acclimation processes of polar condition from Antarctic ice algae Chlamydomonas sp. ICE-L. Mar Biotechnol 13:393–401
Zhong H, Chen J, Li C, Chen L, Wu J, Chen J, Lu W (2011) Selection of reliable reference genes for expression studies by reverse transcription quantitative real-time PCR in litchi under different experimental conditions. Plant Cell Rep 30:641–653
Acknowledgments
This work was supported by Shandong Science and Technology plan project (2011GHY11528), National Natural Science Foundation of China (41176153), National special fund for transgenic project (2009ZX08009-019B), the Hi-Tech Research and Development Program (863) of China (2009AA10Z106), Natural Science Foundation of Shandong Province (2009ZRA02075), Qingdao Municipal Science and Technology plan project (09-2-5-8-hy, 10-4-1-13-hy), National Marine Public Welfare Research Project (200805069) and the National Science & Technology Pillar Program, (2008BAD95B11).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure 1
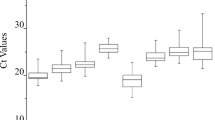
The raw CT values of GAPDH, EF-1α, ACT, H2B, L32 and TUA under different experimental conditions. ■ represents median. The bar indicates the minimal to maximal value. The same letter denotes that there are not significant differences between the genes (p > 0.05). (GIF 44 kb)
Figure 2
The raw CT values of TUB, UBQ, rbcL and 18S under different experimental conditions. ■ represents median. The bar indicates the minimal to maximal value. The same letter denotes that there are not significant differences between the genes (p > 0.05). (GIF 35 kb)
Figure 3
RT-qPCR CT values of the ten candidate reference genes for all tested samples. CT values were inversely proportional to the amount of template.■ represents median. The bar indicates the minimal to maximal value. (GIF 16 kb)
Figure 4
Gene expression stability and ranking of the ten reference genes as calculated by geNorm in all tested samples (A), diurnal cycle (B), HL (C), different light intensity (D), HS (E), and UV-B irradiation (F). A lower average expression stability M value indicated more stable expression. (GIF 169 kb)
Rights and permissions
About this article
Cite this article
Mou, S., Zhang, X., Miao, J. et al. Reference genes for gene expression normalization in Chlamydomonas sp. ICE-L by quantitative real-time RT-PCR. J. Plant Biochem. Biotechnol. 24, 276–282 (2015). https://doi.org/10.1007/s13562-014-0268-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13562-014-0268-4