Abstract
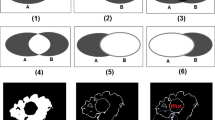
In this paper, we extend our previous work on deformable image registration to inhomogenous tissues. Inhomogenous tissues include the tissues with embedded tumors, which is common in clinical applications. It is a very challenging task since the registration method that works for homogenous tissues may not work well with inhomogenous tissues. The maximum error normally occurs in the regions with tumors and often exceeds the acceptable error threshold. In this paper, we propose a new error correction method with adaptive weighting to reduce the maximum registration error. Our previous fast deformable registration method is used in the inner loop. We have also proposed a new evaluation metric average error of deformation field (AEDF) to evaluate the registration accuracy in regions between vessels and bifurcation points. We have validated the proposed method using liver MR images from human subjects. AEDF results show that the proposed method can greatly reduce the maximum registration errors when compared with the previous method with no adaptive weighting. The proposed method has the potential to be used in clinical applications to reduce registration errors in regions with tumors.






Similar content being viewed by others
References
Huang X, Ren J, Abdalbari A, Green M. Vessel-based fast deformable registration with minimal strain energy. Biomed Eng Lett. 2016;6(1):47–55.
Canadian Cancer Society. http://www.cancer.ca/en/cancer-information/cancer-101/cancer-statistics-at-a-glance/?region=on. Retrieved on 3 Aug 2014.
Ljungberg M, Strand SE. Attenuation and scatter correction in SPECT for sources in a nonhomogeneous object: a Monte Carlo study. J Nucl Med. 1991;32(6):1278–84.
Trivedi A, Basu S, Mitra K. Temporal analysis of reflected optical signals for short pulse laser interaction with nonhomogeneous tissue phantoms. J Quant Spectrosc Radiat Transf. 2005;93(1–3):337–48.
Murphy K, van Ginneken B, Reinhardt JM, Kabus S, Ding K, Deng X, Cao K, Du K, Christensen GE, Garcia V, Vercauteren T, Ayache N, Commowick O, Malandain G, Glocker B, Paragios N, Navab N, Gorbunova V, Sporring J, de Bruijne M, Han X, Heinrich MP, Schnabel JA, Jenkinson M, Lorenz C, Modat M, McClelland JR, Ourselin S, Muenzing SE, Viergever MA, De Nigris D, Collins DL, Arbel T, Peroni M, Li R, Sharp GC, Schmidt-Richberg A, Ehrhardt J, Werner R, Smeets D, Loeckx D, Song G, Tustison N, Avants B, Gee JC, Staring M, Klein S, Stoel BC, Urschler M, Werlberger M, Vandemeulebroucke J, Rit S, Sarrut D, Pluim JP. Evaluation of registration methods on thoracic CT: the EMPIRE10 challenge. IEEE Trans Med Imaging. 2011;30:1901–20.
Han X. Feature-constrained nonlinear registration of lung CT images. Med Image Anal Clinic Grand Chall. 2010;63–72.
Sotiras A, Davatzikos C, Paragios N. Deformable medical image registration: a survey. IEEE Trans Med Imaging. 2013;32(7):1153–9.
Zitova B, Flusser J. Image registration methods: a survey. Image Vis Comput. 2003;21:977–1000.
Hill D, Batchelor P, Holden M, Hawkes D. Medical image registration. Phys Med Biol. 2001;46(3):R1–45.
Xia M, Liu B. Image registration by super curves. IEEE Trans Image Process. 2004;13(5):720–32.
Ren J. Adaptive deformable image registration of in-homogenous tissues. SPIE Med Imagin. 2015.
Baumhauer M, Feuerstein M, Meinzer HP, Rassweiler J. Navigation in endoscopic soft tissue surgery: perspectives and limitations. J Endourol. 2008;22(4):751–66.
Huang X, Bari A, Zaheer S, Looi T, Ren J, Drake J. 3D curve constrained deformable registration using a neuro-fuzzy transformation model. Conf Proc IEEE Eng Med Biol Soc (EMBC2012). 2012;5294–7.
Holden M. A review of geometric transformations for nonrigid body registration. IEEE Trans Med Imaging. 2008;27(1):111–28.
Rueckert D, Sonoda LI, Hayes C, Hill DL, Leach MO, Hawkes DJ. nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans Med Imaging. 1999;18(8):712–21.
Huang Xishi, Ho Danny, Ren Jing, Capretz Luiz F. A soft computing framework for software effort estimation. Soft Comput. 2006;10(2):170–7.
3D Slicer. https://www.slicer.org/.
VMTK. http://www.vmtk.org/.
Noorda YH, Bartels LW, Viergever MA, Pluim JP. Subject-specific four-dimensional liver motion modeling based on registration of dynamic MRI. J Med Imaging. 2016;3(1):015002.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ren, J., Green, M., Huang, X. et al. Automatic error correction using adaptive weighting for vessel-based deformable image registration. Biomed. Eng. Lett. 7, 173–181 (2017). https://doi.org/10.1007/s13534-017-0020-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13534-017-0020-9