Abstract
Using mass-selected ion deposition combined with in situ infrared reflection absorption spectroscopy (IRRAS), we examined the reactive landing of gramicidin S and ubiquitin ions onto activated self-assembled monolayer (SAM) surfaces terminated with N-hydroxysuccinimidyl ester (NHS-SAM) and acyl fluoride (COF-SAM) groups. Doubly protonated gramicidin S, [GS + 2H]2+, and two charge states of ubiquitin, [U + 5H]5+ and [U + 13H]13+, were used as model systems, allowing us to explore the effect of the number of free amino groups and the secondary structure on the efficiency of covalent bond formation between the projectile ion and the surface. For all projectile ions, ion deposition resulted in the depletion of IRRAS bands corresponding to the terminal groups on the SAM and the appearance of several new bands not associated with the deposited species. These new bands were assigned to the C=O stretching vibrations of COOH and COO− groups formed on the surface as a result of ion deposition. The presence of these bands was attributed to an alternative reactive landing pathway that competes with covalent bond formation. This pathway with similar yields for both gramicidin S and ubiquitin ions is analogous to the hydrolysis of the NHS ester bond in solution. The covalent bond formation efficiency increased linearly with the number of free amino groups and was found to be lower for the more compact conformation of ubiquitin compared with the fully unfolded conformation. This observation was attributed to the limited availability of amino groups on the surface of the folded conformation. Our results have provided new insights on the efficiency and mechanism of reactive landing of peptides and proteins onto activated SAMs.

ᅟ
Similar content being viewed by others
Introduction
Preparative mass spectrometry enables precisely controlled physical and chemical modification of solid and liquid materials through collisions of hyperthermal (<150 eV) mass-selected ions with surfaces [1–6]. Physical modification is performed using ion soft-landing, a process where intact projectile ions are deposited onto surfaces with or without charge retention [7]. Ion soft-landing has been used to deposit a wide range of complex molecules, including peptides [8, 9], proteins [10–14], organometallics [15–19], and clusters [20–24]. Meanwhile, chemical modification of surfaces can be achieved using reactive landing, where a bond is formed between the projectile ion and the surface [25–27]. Although many surfaces are well suited for ion deposition experiments, self-assembled monolayers (SAMs) of organic molecules on solid conductive surfaces are popular targets because of their ease of use, reproducibility, and wide selection of terminal functional groups that can be used to control both the reactivity and physical properties of the surface [28]. Reactive landing experiments have been performed on a variety of surfaces [12, 25–27, 29–32]. We are particularly interested in reactive landing of complex multifunctional ions onto activated SAMs terminated by good leaving groups that are substantially more reactive than other SAMs [29, 33, 34]. Reactions of complex gaseous ions, including peptides, dendrimers, and amines with alkylthiol SAMs terminated with N- hydroxysuccinimidyl ester (NHS-SAM) and acyl fluoride (COF-SAM) groups, have been examined experimentally [33–36], whereas reaction dynamics between peptides and SAMs have been explored theoretically using SAMs terminated with aldehyde (CHO-SAM) and acyl chloride (COCl-SAM) groups [37, 38].
Covalent immobilization of peptides and proteins to solid supports is of interest to the development of protein microarray technology [39, 40] synthesis of biocompatible coatings [41, 42], and applications in biosensing [43, 44], among others. Although solution-phase techniques traditionally are used for covalent attachment of proteins to surfaces, the majority of these techniques require relatively large amounts of purified materials that are not always available. In contrast, preparative mass spectrometry can be used for separation and purification of low-abundance species present in complex mixtures, which would greatly simplify the sample preparation stage and enable deposition of high-purity peptide and protein arrays with precise control over the positions of individual spots [11, 45]. We previously demonstrated efficient reactive landing of complex ions containing several reactive functional groups onto NHS-SAM and COF-SAM surfaces [33, 34, 36, 46]. Furthermore, reactive landing of Ac-A15K peptide onto NHS-SAM resulted in covalent immobilization of the α-helical peptide with retention of its stable gas-phase conformation on the surface [45].
NHS-amine chemistry has been extensively used for peptide and protein derivatization both in the gas phase [47–51] and in solution [52–55], as well as for protein immobilization on supports [39, 56–58]. The reaction involves a nucleophilic attack on a carbonyl carbon by a neutral primary amine and, therefore, is pH-dependent in solution. For example, McLuckey and co-workers demonstrated that in solution at pH 5, only the N-terminal amino group of peptide/protein is reactive towards NHS esters, whereas the reactivity of amino groups of lysine side chains is suppressed by protonation [48]. In contrast, lysine side chains are preferentially derivatized using ion–ion reactions between positively charged peptide ions and sulfo-NHS anions in the gas phase [48]. Although the N-terminus was found to be unreactive in positive mode, covalent bond formation was observed in negative mode [48]. This difference in the reactivity of the N-terminal amino group was attributed to its participation in hydrogen bonding interactions affected by the proximity of protonated side chains. Similarly, it has been demonstrated that in reactive landing experiments, the ε-amino group of lysine is involved in covalent attachment of gaseous peptide ions onto NHS-SAM, whereas the N-terminal amino group is unreactive [34]. This conclusion was based on the relative reactive landing yields obtained for several small linear and cyclic peptides with and without lysine residue.
Larger peptide and protein ions typically contain multiple lysine side chains that can participate in covalent bond formation. We previously demonstrated efficient reactive landing of complex multifunctional molecules onto NHS-SAM using several generations of polyamidoamine (PAMAM) dendrimer ions containing multiple amino groups as model systems [36]. We found that reactive landing efficiency increases linearly with an increase in the number of terminal amino groups on the dendrimer surface. In this study, we examined the reactivity of gramicidin S, a cyclic peptide (cyclo-LFPVOLFPVO) that contains two amino groups of the ornithine residues that can react with activated SAMs, and a small protein, ubiquitin, that contains seven reactive lysine residues. We demonstrate that similar to dendrimer ions, the reactive landing efficiency of peptides and proteins is proportional to the number of amino groups. However, unlike dendrimers, reactive landing of peptides and proteins opens up a competing reaction channel that does not result in covalent bond formation between the projectile ion and the surface. Instead, the second channel observed in this study is analogous to the hydrolysis of the NHS ester bond in solution.
Experimental
Preparation of Activated Self-Assembled Monolayer Surfaces
Dithiobis (succinimidyl undecanoate) was purchased from Dojindo Molecular Technologies (Gaithersburg, MD, USA). 16-Mercaptohexadecanoic acid was purchased from Sigma-Aldrich (St. Louis, MO, USA). Cyanuric fluoride (98% packed under argon) was purchased from Alfa Aesar (Ward Hill, MA, USA). Dichloromethane (99%) was purchased from Acros Organics (Morris Plains, NJ, USA). All of the chemicals were used without further purification. SAMs were prepared on 10 × 10-mm silicon wafers with 100-nm gold layer deposited on top of a 10-nm chromium adhesion layer (SPI Supplies, West Chester, PA, USA). Gold-coated surfaces were cleaned in an ultraviolet/ozone cleaner and immediately immersed in 1-mM solutions of the corresponding thiols. NHS-SAM was prepared by immersing a surface in a 1-mM dithiol solution in methylene dichloride:ethanol (20:80 v/v) for 12 h, whereas COOH-SAM was prepared using a 1-mM solution of 16-mercaptohexadecanoic acid in ethanol. The substrates were removed from the thiol solutions, ultrasonically washed in ethanol (NHS-SAM) or 10% v/v acetic acid in ethanol (COOH-SAM) to remove physisorbed molecules from the SAM surface, and dried under a nitrogen stream. The acyl fluoride-terminated SAM (COF-SAM) was prepared via derivatization of the COOH-SAM using a literature procedure [59]. Namely, the COOH-SAM was immersed in a mixture of 4 μL cyanuric acid, 100 μL pyridine, and 10 mL dichloromethane for 1 h at room temperature, ultrasonically washed in ethanol, and dried under a nitrogen stream. This procedure is known to result in complete derivatization of COOH terminal groups on a SAM surface [59].
Reactive Landing of Peptide and Protein Ions
Gramicidin S and bovine ubiquitin were purchased from Sigma-Aldrich (St. Louis, MO, USA) and used as received. [GS + 2H]2+ ions were produced by electrospray ionization (ESI) of a 100-μM solution of gramicidin S in methanol. Ubiquitin ions were produced by ESI of a 50-μM solution. The lower 5+ charge state of ubiquitin, [U + 5H]5+, was produced from a mixture of H2O:CH3OH:NH4HCO3 (49:49:2 v/v/v), whereas the higher 13+ charge state, [U + 13H]13+, was produced using a CH3OH:H2O:CH3COOH:glycerol (48.5:48.5:2:1 v/v/v) mixture as a solvent [60]. Mass-selected [GS + 2H]2+, [U + 5H]5+, and [U + 13H]13+ were used as projectile ions in reactive landing experiments. Typical ion currents measured on the surface using a Keithley Instruments (Solon, OH, USA) electrometer were as follows: 400 pA for [GS + 2H]2+, 40 pA for [U + 5H]5+, and 180 pA for [U + 13H]13+.
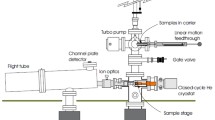
Reactive landing experiments were performed using the ion deposition apparatus described in detail elsewhere [35, 61]. This instrument enables in situ IRRAS characterization of surfaces during and after ion soft-landing. Charged droplets produced in an ESI source undergo desolvation in a heated inlet maintained at ~100 °C to generate positively charged ions of analyte molecules. Ions are transferred into vacuum through an electrodynamic ion funnel, focused in a collisional quadrupole (CQ), and mass selected using a commercial quadrupole mass filter (Extrel, Pittsburgh, PA, USA). Mass-selected ions exiting the mass filter are refocused and positioned using two Einzel lenses before colliding with the surface located in a vacuum chamber maintained at 6 × 10−5 Torr. The nominal collision energy per charge of the ion, defined as the potential difference between the direct current offset of CQ and the surface, was kept at ~30 eV. Our previous study demonstrated that for multiply charged dendrimer ions, the reactive landing efficiency is only weakly dependent on the collision energy of the projectile ion [36]. The number of deposited ions was estimated based on the ion current measured on the deposition target assuming that all ions are retained on the surface.
In Situ Surface Characterization Using Infrared Reflection Absorption Spectroscopy
Grazing-incidence IRRAS experiments were performed using a Bruker Vertex 70 Fourier transform infrared (FTIR) spectrometer (Bruker Optics, Billerica, MA, USA) equipped with a liquid nitrogen-cooled mercury cadmium telluride (MCT) detector. The experimental platform is described in detail in our previous study [35]. Briefly, IRRAS spectra are obtained by directing infrared light onto the SAM sample positioned inside the vacuum chamber. Infrared light exiting the spectrometer is deflected using a flat, gold-coated mirror and focused using a 90° gold-coated parabolic mirror with a 50-mm diameter and focal length of 400 mm. The light then passes the mid-infrared (MIR) KRS-5 wire grid polarizer and is transferred into the vacuum system through the wedged ZnSe vacuum viewport (Laser Optex Inc., Beijing, China). The surface is positioned in the focal point of the parabolic mirror. Reflected light leaves the vacuum system through the second ZnSe viewport and is focused onto the MCT detector using the second parabolic mirror. In this study, spectra were obtained at a resolution of 4 cm−1 using p-polarized light. The angle of incidence was 80° with respect to the surface normal. The infrared beam path was purged with nitrogen gas. IRRAS spectra were recorded every 10 min by averaging 512 scans corresponding to an acquisition time of 2 min/spectrum. Water subtraction and baseline correction were performed using OPUS software.
Fitting the IRRAS spectra was performed using Excel. Specifically, spectral data in the 1000–2200 cm−1 region were reproduced with a sum of nine Lorentzian curves providing the positions and intensities of the major bands observed in this region of the IRRAS spectrum. All time-resolved IRRAS data obtained in one experiment were simultaneously analyzed. The best fit was obtained by minimizing sums of squares of deviations between the experimental and simulated curves in Excel’s Solver using the generalized reduced gradient (GRG) nonlinear algorithm. Fitting involved iterative optimization of the band positions, widths, and intensities of individual components until the best fit was obtained, assuming both the position and width of each band did not change as a function of time.
Results and Discussion
In this study, we examined reactive landing of the doubly protonated gramicidin S, [GS + 2H]2+, and two charge states of ubiquitin, [U + 5H]5+ and [U + 13H]13+, onto activated NHS-SAM and COF-SAM surfaces. Our previous studies demonstrated efficient covalent attachment of peptide ions to NHS-SAM via an unprotonated ε-amino group of a lysine side chain and only a minor yield of reactive landing through an N-terminal amino group [33, 34]. Furthermore, we found no evidence of peptide binding to NHS-SAM through other side chains. Efficient reactive landing also has been reported for PAMAM dendrimer ions decorated with free amino groups [36]. Interestingly, for dendrimer ions, the reactive landing efficiency increased in proportion to the number of terminal functional groups on the surface. Peptide and protein ions often contain multiple amino groups available for reactions with SAM surfaces. However, these groups may not be available for reaction because of protonation, hydrogen bonding interactions, and the higher-order (secondary, tertiary) structure of the ion. Model systems used in this study, gramicidin S and ubiquitin, allowed us to examine the effect of the number of amino groups and protein conformation on reactive landing efficiency.
Two charge states of ubiquitin, 5+ and 13+, were selected to represent compact and elongated conformations, respectively [62–64]. Previously, we demonstrated that the initial charge state of ubiquitin has a measurable effect on its conformation on the surface with a higher abundance of β-sheet and helical conformations observed for the respective 5+ and 13+ charge states [60]. Furthermore, we found that for both peptide and dendrimer ions, the reactive landing efficiency is independent of the initial charge state [33, 34, 36]. This observation was attributed to neutralization of projectile ions during collision with NHS-SAM. Similar to peptide and dendrimer ions, assuming protein ions undergo partial neutralization during deposition, we hypothesize that exposure of lysine side chains controlled by the secondary structure of deposited ubiquitin ions is a major factor that may have a pronounced effect on reactive landing efficiency. Specifically, it is reasonable to assume that in the elongated conformation of the 13+ charge state, lysine residues are fully exposed and available for reactive landing. Meanwhile, lysine residues are partially shielded in the more compact native-like conformation of the 5+ charge state [65]. Differences in the neutralization efficiency of the 5+ and 13+ charge states of ubiquitin ions will have an opposing effect on reactive landing efficiency with the higher charge state expected to be less reactive than the lower charge state.
The efficiency and mechanisms of reactive landing were explored using in situ IRRAS characterization of surfaces during ion deposition. In these experiments, mass-selected [GS + 2H]2+, [U + 5H]5+, and [U + 13H]13+ ions were deposited onto two different activated SAMs (NHS-SAM and COF-SAM). Time-resolved IRRAS spectra were acquired at regular intervals during ion deposition. The kinetic energy of the ions was 30 eV/charge corresponding to the nominal kinetic energy of 60 eV, 150 eV, and 390 eV for the 2+, 5+, and 13+ ions, respectively. Of note is that time-resolved IRRAS experiments are always conducted with the surface at the ground potential and ion’s nominal kinetic energy is determined by the offset of the collisional quadrupole. Different ion optics without optical access to the surface is used when biasing the surface off the ground potential is necessary. High nominal kinetic energies used in this study are typical for reactive landing of protein ions [12]. Although the ion’s kinetic energy may affect reactive landing efficiency [25, 34], our previous study demonstrated that for both singly and doubly charged peptide ions the nominal kinetic energy of ~30 eV is sufficient to overcome the reaction barrier on NHS-SAM, and the reactive landing efficiency remains constant over a fairly broad range of kinetic energies [34]. In contrast with peptide ions that showed measurable decline of the reactive landing efficiency at higher kinetic energies (>80 eV) [34], similar reactive landing efficiency was obtained for dendrimer ions in charge states in a range of 1 to 7 deposited onto NHS-SAM at nominal collision energies in a range of 30 to 120 eV [36]. It is reasonable to assume that for the relatively high kinetic energy and size of projectile ions used in this study, the reactive landing efficiency of peptide and protein ions also is independent of ion’s collision energy. Chemical dynamics simulations could be used to test this hypothesis [29, 66].
Typical IRRAS spectra of [GS + 2H]2+, [U + 5H]5+, and [U + 13H]13+ on NHS-SAM are shown in Figure 1, and time-resolved IRRAS spectra are shown in Supplementary Figure S1. IRRAS spectra reported in this study were acquired using spectra of unmodified SAMs obtained prior to each ion deposition as a background. Negative signals observed in the spectra correspond to the depletion of the characteristic NHS-SAM bands following ion deposition. Specifically, the bands corresponding to the stretching vibration of the succinimidyl carbonyl groups at 1752 cm−1 and the asymmetric CNC stretching of the NHS at 1221 cm−1 are suppressed during the reactive landing experiment [34–36]. Positive signals correspond to the characteristic amide I, amide II, and amide III vibrations of gramicidin S and ubiquitin. Another band around 1740 cm−1, marked with an asterisk in Figure 1, is observed for both species as a shoulder on the negative C=O stretching band of the NHS group. This band is absent in the spectra of diamine and smaller dendrimer ions but is clearly present at lower abundance in the spectrum of the 4+ charge state of the generation 2 PAMAM dendrimer shown in Figure 1 for comparison.
Representative IRRAS spectra obtained after reactive landing of [GS + 2H]2+ (red), [U + 5H]5+ (green), and [U + 13H]13+ (blue) ions onto an NHS-SAM surface. The IRRAS spectrum of the 4+ charge state of the generation 2 PAMAM dendrimer, [DEN + 4H]4+, is shown for comparison (black). The unmodified NHS-SAM was used as a background. Several bands highlighted in the figure include the stretching vibration of the succinimidyl carbonyl groups at 1752 cm−1 and the asymmetric CNC stretching of the NHS at 1220 cm−1 depleted due to reactive landing of ions along with the amide I, II, and III bands of the deposited species. The newly formed band around 1740 cm−1 marked with an asterisk (*) is discussed in detail in the text
To understand the origin of this band, we performed reactive landing experiments using COF-SAM as a deposition target. The C=O stretch of this activated SAM is shifted to 1831 cm−1 and does not overlap with the unassigned band [35]. Figure 2 shows the IRRAS spectra obtained after deposition of [GS + 2H]2+ and [U + 5H]5+ on COF-SAM, along with the spectrum of the unmodified surface. Similar to NHS-SAM, we observe the depletion of the C=O stretch of COF-SAM due to ion deposition and the growth of the characteristic amide vibrational bands. Another new band at 1713 cm−1 appears in the spectrum of both gramicidin S and ubiquitin on COF-SAM as a shoulder of the amide I band. The origin of this band is discussed in detail later herein.
IRRAS spectrum of the COF-SAM surface obtained using clean gold surface as a background along with IRRAS spectra of the COF-SAM after deposition of [GS + 2H]2+ and [U + 5H]5+ obtained using unmodified COF-SAM as a background. Bands highlighted in the figure include the stretching C=O vibration of the acyl fluoride at 1841 cm−1 depleted due to reactive landing of ions along with the amide I, II, and III bands of the deposited species. The newly formed band around 1720 cm−1 marked with an asterisk (*) is discussed in detail in the text
Fitting the time-resolved IRRAS data across the 1000–2200 cm−1 range using a sum of nine Lorentzian functions (basis functions) was performed as described in detail in the Experimental section. For NHS-SAM, two bands corresponding to the C=O (1752 cm−1) and CNC (1221 cm−1) stretching vibrations of the NHS group were assigned negative intensities while the intensities of the remaining seven bands were kept at positive values. Figure 3 depicts an example of the simulated curve obtained for [GS + 2H]2+. Table 1 summarizes the band positions and relative intensities averaged over all of the time-resolved IRRAS spectra obtained on NHS-SAM. The IRRAS signal in the 1710–1760 cm−1 range was reproduced using three basis functions. The shape of the amide I band was adequately reproduced using two basis functions, whereas only one basis function was used for each of the remaining bands. For all three projectile ions, the dominant band in the IRRAS spectra corresponds to the stretching vibration of the NHS carbonyl groups at 1752 cm−1. Consistent with our previous work, two bands corresponding to the α-helix (1663 cm−1) and parallel β-sheet (1676 cm−1) are the most abundant features of the signal observed in the amide I region of [U + 5H]5+ and [U + 13H]13+ [60]. Meanwhile, the amide I band of the soft-landed [GS + 2H]2+ is centered around 1663 cm−1. The band’s position is consistent with the helical or random coil conformation of gramicidin S on NHS-SAM. The position of the amide III band of gramicidin S (1303 cm−1) is consistent with the helical conformation. However, the amide III band in the IRRAS spectrum of [GS + 2H]2+ on NHS-SAM is broader than the corresponding band of the α-helical Ac-A15K peptide on the same surface [45], indicating that several secondary structure motifs contribute to the observed IRRAS signal. The presence of helical conformations of gramicidin S on NHS-SAM is unexpected as this peptide is known to assume an antiparallel β-sheet conformation in the condensed phase [67]. The shape of the amide I band in the IRRAS spectrum of soft-landed [GS + 2H]2+ is also quite different from the infrared spectrum reported for this ion in the gas phase [68], suggesting that the secondary structure of deposited gramicidin S is different from its gas-phase structure. Finally, we note that the position of the amide I band of gramicidin S on NHS-SAM is blue-shifted by more than 10 cm−1 in comparison with FSAM [35], suggesting that interactions with the NHS-SAM surface have a pronounced effect on the conformation of this peptide. Detailed understanding of the effect of the surface on the secondary structure of gramicidin S is outside the scope of this manuscript. Meanwhile, the major amide I components of ubiquitin on NHS-SAM are consistent with those observed in our previous study [60]. Furthermore, the peak at ~1270 cm−1 in the amide III region of the ubiquitin spectra most likely corresponds to random coils or β-turns [69, 70] observed in our previous study for ubiquitin ions soft-landed onto COOH-SAM [60].
In addition to the amide bands of the deposited peptide and protein ions, three new bands positioned around 1745 cm−1, 1729 cm−1, and 1457 cm−1 are clearly distinguished using curve fitting. For example, fitting the experimental data allowed us to separate the newly formed 1745 cm−1 and 1729 cm−1 from the negative NHS band at 1752 cm−1 (shown in Figure 3). The location of the 1745 cm−1 and 1729 cm−1 bands is consistent with the stretching vibration of the non-hydrogen-bonded and hydrogen-bonded carbonyl groups of COOH, respectively [71]. The 1457 cm−1 band may be attributed to the carboxylate (COO−) group [71, 72]. Notably, the relative abundance of these three bands is similar for all projectile ions, indicating that the formation of these bands is not sensitive to the size, composition, and conformation of peptide or protein species. Although acidic side chains could contribute to the presence of these bands, their contribution to the spectrum of [GS + 2H]2+ can be ruled out because gramicidin S does not contain carboxyl groups. Similarly, covalent bond formation through reactions of other side chains (e.g., serine or arginine) with NHS-SAM can be ruled out due to the absence of any reactive side chains in gramicidin S. The appearance of the 1745 cm−1 band also could be attributed to the red shift in the position of the 1752 cm−1 band of the NHS group stemming from noncovalent interactions with the deposited species. Based on the preceding discussion, we propose that detachment of the NHS group due to reactive landing of gramicidin S and ubiquitin is not always followed by covalent bond formation between the projectile and the surface. Instead, a fraction of collisions results in a process analogous to hydrolysis of the NHS ester bond in solution. Furthermore, the presence of COO− groups indicates partial proton transfer from the newly formed COOH groups to the deposited species. The efficiency of ester hydrolysis during reactive landing of peptides and proteins is surprisingly high in comparison with the efficiency observed for dendrimer ions (Figure 1). Thus, based on our discussion, it follows that the observed depletion of the NHS band at 1752 cm−1 results from at least two competing processes: ester hydrolysis and covalent bond formation.
In comparison with NHS-SAM, the band at 1745 cm−1 is not observed on COF-SAM (Figure 2). Two new bands at 1713 cm−1 and 1457 cm−1 observed for both [GS + 2H]2+ and [U + 5H]5+ deposited onto COF-SAM most likely correspond to the hydrogen-bonded carbonyl stretching and carboxylate band, respectively. The relative abundance of the band at 1713 cm−1 is ~25% of the major amide I band at 1668 cm−1. Meanwhile, the relative abundance of the carboxylate band formed on this surface is ~34% of the major amide I at 1668 cm−1. These values are comparable to the relative abundances of the 1729 cm−1 and 1457 cm−1 bands observed using NHS-SAM as a deposition target. We propose that the band at 1713 cm−1 corresponds to the hydrogen-bonded carbonyl stretching band. The shift in this band’s position from 1729 cm−1 for NHS-SAM to 1713 cm−1 for COF-SAM indicates stronger hydrogen bonding interactions between COOH groups formed on COF-SAM during reactive landing experiments compared with NHS-SAM. The absence of the 1745 cm−1 band in the spectra of gramicidin S and ubiquitin on COF-SAM could be attributed either to the unfavorable orientation of the newly formed COOH groups or to the absence of the non-hydrogen-bonded COOH groups on this surface. Alternatively, it could indicate that the 1745 cm−1 band on the NHS-SAM surface originates from the red shift in the position of the 1752 cm−1 NHS band due to interactions between the deposited species and the surface. Regardless of the band’s origin, the presence of two other bands characteristic of the carboxyl group on both NHS-SAM and COF-SAM surfaces is consistent with the alternative mechanism of reactive landing that does not result in the covalent bond formation between the projectile ion and the surface. Interestingly, related infrared bands have been reported for polylysine adsorbed onto NHS-SAM from solution [73].
In our previous study, the reaction extent was estimated based on depletion of the NHS band at 1753 cm−1 relative to the absorbance of this band prior to ion deposition [36]. Figure 4 shows the dependence of NHS band depletion on the number of deposited [GS + 2H]2+, [U + 5H]5+, and [U + 13H]13+ ions. Previously reported data obtained for the generation 0 (DEN G0, [M + H]+) and generation 3 (DEN G3, [M + 7H]7+) dendrimer ions are included for comparison. Similar to previously published data, the NHS band depletion is a linear function of the number of deposited ions [35, 36]. However, the NHS band depletion observed in this study is more efficient compared with the data obtained for dendrimer ions. For example, although gramicidin S contains two amino groups available for reaction, it shows a similar rate of NHS band depletion as DEN G0 that contains four amino groups. Furthermore, the NHS band depletion observed for ubiquitin (seven amino groups) is similar to the depletion reported for DEN G3 (32 amino groups). As discussed earlier, two competing processes contribute to the NHS band depletion in reactive landing of peptide and protein ions. The efficiency of the covalent bond formation was estimated by subtracting the absorbance of the dominant band at 1745 cm−1 from the absolute absorbance of the 1753 cm−1 band. The relative reaction efficiency is given by the slope of the linear plot of the absolute absorbance of the NHS band corrected for the contribution of the major COOH band. Figure 5 shows the dependence of the relative reaction efficiency on the number of amino groups available for reaction. The results obtained in this study for the covalent attachment of gramicidin S and ubiquitin to NHS-SAM are consistent with previously published data [36]. Specifically, the reaction efficiency shows a linear increase with increases in the number of NH2 groups, indicating that similar to dendrimers, reactive landing of peptides and proteins results in the formation of multiple amide bonds with NHS-SAM. Of note, there is a measurable difference in the covalent binding efficiency of [U + 5H]5+ and [U + 13H]13+ ions, which most likely can be attributed to the limited availability of amino groups on the surface of the more compact conformation of [U + 5H]5+ compared with the extended conformation of [U + 13H]13+ [65].
The depletion of the NHS band at 1753 cm−1 as a function of the number of deposited [GS + 2H]2+ (black squares), [U + 5H]5+ (red circles), and [U + 13H]13+ (blue triangles) ions. For comparison, depletion of the NHS band observed for the [M + H]+ ion of the generation 0 PAMAM dendrimer (DEN G0, dashed line) and [M + 7H]7+ of the generation 3 PAMAM dendrimer (DEN G3, solid line) also are shown
Conclusions
Reactive landing of gramicidin S and ubiquitin ions onto activated SAMs terminated with labile NHS and COF groups was studied using mass-selected ion deposition coupled with surface characterization by time-resolved in situ IRRAS. Similar to our previous studies, ion deposition resulted in the depletion of the carbonyl stretching bands of NHS-SAM and COF-SAM surfaces. For dendrimer ions, this depletion was attributed to the formation of a covalent bond between the projectile ion and the surface. However, for gramicidin S and ubiquitin examined in this study, the observed depletion of the characteristic surface bands was attributed to the competition between the covalent bond formation and displacement of the SAM’s labile end group. The latter process results in the formation of new bands corresponding to the C=O stretching vibrations of COOH and COO− and resembles ester hydrolysis in solution. These observations can be rationalized assuming that the reaction occurs in a stepwise manner. The first step involves cleavage of the NHS ester bond that most likely occurs during collision, whereas the second step involves either the covalent bond formation or loss of a water molecule from the deposited species to the surface. The efficiency of covalent bond formation observed for gramicidin S and ubiquitin follows the same linear increase with the number of available amino groups as the one reported for dendrimer ions. We also observed the effect of the protein conformation on reactive landing efficiency. Specifically, lower covalent bond formation efficiency was observed for the more compact conformation of ubiquitin, in which some lysine side chains are protected from the reactive group by the secondary structure and hydrogen bonding interactions. These results have provided new insights on key processes that occur during reactive landing of peptides and proteins onto activated SAMs. Chemical dynamics simulations highlighted in recent reviews could provide additional insights on the structures of reactively landed peptides and proteins along with ester hydrolysis at the time of collision [29, 66].
References
Grill, V., Shen, J., Evans, C., Cooks, R.G.: Collisions of ions with surfaces at chemically relevant energies: instrumentation and phenomena. Rev. Sci. Instrum. 72, 3149–3179 (2001)
Gologan, B., Green, J.R., Alvarez, J., Laskin, J., Cooks, R.G.: Ion/surface reactions and ion soft-landing. PCCP 7, 1490–1500 (2005)
Johnson, G.E., Hu, Q.C., Laskin, J.: Soft landing of complex molecules on surfaces. Annu. Rev. Anal. Chem. 4, 83–104 (2011)
Cyriac, J., Pradeep, T., Kang, H., Souda, R., Cooks, R.G.: Low-energy ionic collisions at molecular solids. Chem. Rev. 112, 5356–5411 (2012)
Verbeck, G., Hoffmann, W., Walton, B.: Soft-landing preparative mass spectrometry. Analyst 137, 4393–4407 (2012)
Johnson, G.E., Gunaratne, D., Laskin, J.: Soft- and reactive landing of ions onto surfaces: concepts and applications. Mass Spectrom. Rev. 35, 439–479 (2016)
Miller, S.A., Luo, H., Pachuta, S.J., Cooks, R.G.: Soft-landing of polyatomic ions at fluorinated self-assembled monolayer surfaces. Science 275, 1447 (1997)
Alvarez, J., Futrell, J.H., Laskin, J.: Soft-landing of peptides onto self-assembled monolayer surfaces. J. Phys. Chem. A 110, 1678–1687 (2006)
Laskin, J., Wang, P., Hadjar, O.: Soft-landing of peptide ions onto self-assembled monolayer surfaces: an overview. PCCP 10, 1079–1090 (2008)
Gologan, B., Takats, Z., Alvarez, J., Wiseman, J.M., Talaty, N., Ouyang, Z., Cooks, R.G.: Ion soft-landing into liquids: protein identification, separation, and purification with retention of biological activity. J. Am. Soc. Mass Spectrom. 15, 1874–1884 (2004)
Ouyang, Z., Takats, Z., Blake, T.A., Gologan, B., Guymon, A.J., Wiseman, J.M., Oliver, J.C., Davisson, V.J., Cooks, R.G.: Preparing protein microarrays by soft-landing of mass-selected ions. Science 301, 1351–1354 (2003)
Volny, M., Elam, W.T., Branca, A., Ratner, B.D., Turecek, F.: Preparative soft and reactive landing of multiply charged protein ions on a plasma-treated metal surface. Anal. Chem. 77, 4890–4896 (2005)
Deng, Z.T., Thontasen, N., Malinowski, N., Rinke, G., Harnau, L., Rauschenbach, S., Kern, K.: A close look at proteins: submolecular resolution of two- and three-dimensionally folded cytochrome c at surfaces. Nano Lett. 12, 2452–2458 (2012)
Rinke, G., Rauschenbach, S., Harnau, L., Albarghash, A., Pauly, M., Kern, K.: Active conformation control of unfolded proteins by hyperthermal collision with a metal surface. Nano Lett. 14, 5609–5615 (2014)
Judai, K., Sera, K., Amatsutsumi, S., Yagi, K., Yasuike, T., Yabushita, S., Nakajima, A., Kaya, K.: A soft-landing experiment on organometallic cluster ions: infrared spectroscopy of V(benzene)(2) in Ar matrix. Chem. Phys. Lett. 334, 277–284 (2001)
Nagaoka, S., Matsumoto, T., Okada, E., Mitsui, M., Nakajima, A.: Room-temperature isolation of V(benzene)(2) sandwich clusters via soft-landing into N-alkanethiol self-assembled monolayers. J. Phys. Chem. B 110, 16008–16017 (2006)
Nagaoka, S., Ikemoto, K., Horiuchi, K., Nakajima, A.: Soft- and reactive-landing of Cr(aniline)(2) sandwich complexes onto self-assembled monolayers: separation between functional and binding sites. J. Am. Chem. Soc. 133, 18719–18727 (2011)
Johnson, G.E., Laskin, J.: Preparation of surface organometallic catalysts by gas-phase ligand stripping and reactive landing of mass-selected ions. Chem. Eur. J. 16, 14433–14438 (2010)
Cooks, R.G., Peng, W.P., Johnson, G.E., Fortmeyer, I.C., Wang, P., Hadjar, O., Laskin, J.: Redox chemistry in thin layers of organometallic complexes prepared using ion soft landing. PCCP 13, 267–275 (2011)
Tyo, E.C., Vajda, S.: Catalysis by clusters with precise numbers of atoms. Nat. Nanotechnol. 10, 577–588 (2015)
Johnson, G.E., Wang, C., Priest, T., Laskin, J.: Monodisperse Au11 clusters prepared by soft landing of mass selected ions. Anal. Chem. 83, 8069–8072 (2011)
Heiz, U., Bullock, E.L.: Fundamental aspects of catalysis on supported metal clusters. J. Mater. Chem. 14, 564–577 (2004)
Popok, V.N., Barke, I., Campbell, E.E.B., Meiwes-Broer, K.H.: Cluster-surface interaction: from soft landing to implantation. Surf. Sci. Rep. 66, 347–377 (2011)
Kahle, S., Deng, Z.T., Malinowski, N., Tonnoir, C., Forment-Aliaga, A., Thontasen, N., Rinke, G., Le, D., Turkowski, V., Rahman, T.S., Rauschenbach, S., Ternes, M., Kern, K.: The quantum magnetism of individual manganese-12-acetate molecular magnets anchored at surfaces. Nano Lett. 12, 518–521 (2012)
Shen, J.W., Grill, V., Evans, C., Cooks, R.G.: Chemical modification of fluorinated self-assembled monolayer surfaces using low-energy ion beams for halogen and pseudohalogen transfer. J. Mass Spectrom. 34, 354–363 (1999)
Wade, N., Gologan, B., Vincze, A., Cooks, R.G., Sullivan, D.M., Bruening, M.L.: Esterification and ether formation at a hydroxyl-terminated self-assembled monolayer surface using low-energy collisions of polyatomic cations. Langmuir 18, 4799–4808 (2002)
Wang, P., Laskin, J.: Surface modification using reactive landing of mass-selected ions. In: Hellborg, R., Whitlow, H.J., Zhang, Y. (eds.) Ion Beams in Nanoscience and Technology, pp. 37–65. Springer-Verlag, Berlin (2009)
Cooks, R.G., Ast, T., Pradeep, T., Wysocki, V.: Reactions of ions with organic surfaces. Acc. Chem. Res. 27, 316–323 (1994)
Pratihar, S., Barnes, G.L., Laskin, J., Hase, W.L.: Dynamics of protonated peptide ion collisions with organic surfaces: consonance of simulation and experiment. J. Phys. Chem. Lett. 7, 3142–3150 (2016)
Blacken, G.R., Volny, M., Vaisar, T., Sadılek, M., Turecek, F.: In situ enrichment of phosphopeptides on MALDI plates functionalized by reactive landing of zirconium(Iv)-N-propoxide ions. Anal. Chem. 79, 5449–5456 (2007)
Mazzei, F., Favero, G., Frasconi, M., Tata, A., Pepi, F.: Electron-transfer kinetics of microperoxidase-11 covalently immobilised onto the surface of multi-walled carbon nanotubes by reactive landing of mass-selected ions. Chem. Eur. J. 15, 7359–7367 (2009)
Pepi, F., Tata, A., Garzoli, S., Giacomello, P., Ragno, R., Patsilinakos, A., Di Fusco, M., D'Annibale, A., Cannistraro, S., Baldacchini, C., Favero, G., Frasconi, M., Mazzei, F.: Chemically modified multiwalled carbon nanotubes electrodes with ferrocene derivatives through reactive landing. J. Phys. Chem. C 115, 4863–4871 (2011)
Wang, P., Hadjar, O., Laskin, J.: Covalent immobilization of peptides on self-assembled monolayer surfaces using soft-landing of mass-selected ions. J. Am. Chem. Soc. 129, 8682–8683 (2007)
Wang, P., Hadjar, O., Gassman, P.L., Laskin, J.: Reactive landing of peptide ions on self-assembled monolayer surfaces: an alternative approach for covalent immobilization of peptides on surfaces. PCCP 10, 1512–1522 (2008)
Hu, Q.C., Wang, P., Gassman, P.L., Laskin, J.: In situ studies of soft- and reactive landing of mass-selected ions using infrared reflection absorption spectroscopy. Anal. Chem. 81, 7302–7308 (2009)
Hu, Q., Laskin, J.: Reactive landing of dendrimer ions onto activated self-assembled monolayer surfaces. J. Phys. Chem. C 118, 2602–2608 (2014)
Barnes, G.L., Young, K., Yang, L., Hase, W.L.: Fragmentation and reactivity in collisions of protonated diglycine with chemically modified perfluorinated alkylthiolate-self-assembled monolayer surfaces. J. Chem. Phys. 134, 094106 (2011)
Geragotelis, A., Barnes, G.L.: Surface deposition resulting from collisions between diglycine and chemically modified alkylthiolate self-assembled monolayer surfaces. J. Phys. Chem. C 117, 13087–13093 (2013)
Rusmini, F., Zhong, Z., Feijen, J.: Protein immobilization strategies for protein biochips. Biomacromolecules 8, 1775–1789 (2007)
Brady, D., Jordaan, J.: Advances in enzyme immobilization. Biotechnol. Lett. 31, 1639–1650 (2009)
Costa, F., Carvalho, I.F., Montelaro, R.C., Gomes, P., Martins, M.C.L.: Covalent immobilization of antimicrobial peptides (Amps) onto biomaterial surfaces. Acta Biomater. 7, 1431–1440 (2011)
Glinel, K., Thebault, P., Humblot, V., Pradier, C.M., Jouenne, T.: Antibacterial surfaces developed from bio-inspired approaches. Acta Biomater. 8, 1670–1684 (2012)
Williams, R.A., Blanch, H.W.: Covalent immobilization of protein monolayers for biosensor applications. Biosens. Bioelectron. 9, 159–167 (1994)
Cosnier, S.: Biomolecule immobilization on electrode surfaces by entrapment or attachment to electrochemically polymerized films. A review. Biosens. Bioelectron. 14, 443–456 (1999)
Wang, P., Laskin, J.: Helical peptide arrays on self-assembled monolayer surfaces through soft and reactive landing of mass-selected ions. Angew. Chem. Int. Ed. 47, 6678–6680 (2008)
Johnson, G.E., Lysonski, M., Laskin, J.: In situ reactivity and TOF-SIMS analysis of surfaces prepared by soft and reactive landing of mass-selected ions. Anal. Chem. 82, 5718–5727 (2010)
Mentinova, M., McLuckey, S.A.: Covalent modification of gaseous peptide ions with n-hydroxysuccinimide ester reagent ions. J. Am. Chem. Soc. 132, 18248–18257 (2010)
Mentinova, M., Barefoot, N.Z., McLuckey, S.A.: Solution versus gas-phase modification of peptide cations with nhs-ester reagents. J. Am. Soc. Mass Spectrom. 23, 282–289 (2012)
Webb, I.K., Mentinova, M., McGee, W.M., McLuckey, S.A.: Gas-phase intramolecular protein crosslinking via ion/ion reactions: ubiquitin and a homobifunctional sulfo-NHS ester. J. Am. Soc. Mass Spectrom. 24, 733–743 (2013)
Peng, Z., McGee, W.M., Bu, J.X., Barefoot, N.Z., McLuckey, S.A.: Gas-phase reactivity of carboxylates with N-hydroxysuccinimide esters. J. Am. Soc. Mass Spectrom. 26, 174–180 (2015)
Bu, J.X., Fisher, C.M., Gilbert, J.D., Prentice, B.M., McLuckey, S.A.: Selective covalent chemistry via gas-phase ion/ion reactions: an exploration of the energy surfaces associated with N-hydroxysuccinimide ester reagents and primary amines and guanidine groups. J. Am. Soc. Mass Spectrom. 27, 1089–1098 (2016)
Elia, G.: Biotinylation reagents for the study of cell surface proteins. Proteomics 8, 4012–4024 (2008)
Roberts, M.J., Bentley, M.D., Harris, J.M.: Chemistry for peptide and protein pegylation. Adv Drug Deliv. Rev. 64(Supplement), 116–127 (2012)
Sinz, A.: Chemical cross-linking and mass spectrometry for mapping three-dimensional structures of proteins and protein complexes. J. Mass Spectrom. 38, 1225–1237 (2003)
Mattson, G., Conklin, E., Desai, S., Nielander, G., Savage, M.D., Morgensen, S.: A practical approach to crosslinking. Mol. Biol. Rep. 17, 167–183 (1993)
MacBeath, G., Schreiber, S.L.: Printing proteins as microarrays for high-throughput function determination. Science 289, 1760–1763 (2000)
Lim, C.Y., Owens, N.A., Wampler, R.D., Ying, Y., Granger, J.H., Porter, M.D., Takahashi, M., Shimazu, K.: Succinimidyl ester surface chemistry: implications of the competition between aminolysis and hydrolysis on covalent protein immobilization. Langmuir 30, 12868–12878 (2014)
Frey, B.L., Corn, R.M.: Covalent attachment and derivatization of poly(L-lysine) monolayers on gold surfaces as characterized by polarization − modulation FT-IR spectroscopy. Anal. Chem. 68, 3187–3193 (1996)
Chi, Y.S., Choi, I.S.: Reactivity control of carboxylic acid-terminated self-assembled monolayers on gold: acid fluoride versus interchain carboxylic anhydride. Langmuir 21, 11765–11772 (2005)
Hu, Q., Laskin, J.: Secondary structures of ubiquitin ions soft-landed onto self-assembled monolayer surfaces. J. Phys. Chem. B 120, 4927–4936 (2016)
Laskin, J., Hadjar, O., Wang, P., Futrell, J.H., Dessiaterik, Y., Zhu, Z.H., Cowin, J.P., Iedema, M.J.: Design and performance of an instrument for soft landing of biomolecular ions on surfaces. Anal. Chem. 79, 6566–6574 (2007)
Goldstein, M., Zmiri, L., Segev, E., Wyttenbach, T., Gerber, R.B.: An atomistic structure of ubiquitin + 13 relevant in mass spectrometry: theoretical prediction and comparison with experimental cross-sections. Int. J. Mass Spectrom. 367, 10–15 (2014)
Purves, R.W., Barnett, D.A., Ells, B., Guevremont, R.: Elongated conformers of charge states +11 to +15 of bovine ubiquitin studied using ESI-FAIMS-MS. J. Am. Soc. Mass Spectrom. 12, 894–901 (2001)
Breuker, K., Oh, H.B., Horn, D.M., Cerda, B.A., McLafferty, F.W.: Detailed unfolding and folding of gaseous ubiquitin ions characterized by electron capture dissociation. J. Am. Chem. Soc. 124, 6407–6420 (2002)
Novak, P., Kruppa, G.H., Young, M.M., Schoeniger, J.: A top-down method for the determination of residue-specific solvent accessibility in proteins. J. Mass Spectrom. 39, 322–328 (2004)
Pratihar, S., Barnes, G.L., Hase, W.L.: Chemical dynamics simulations of energy transfer, surface-induced dissociation, soft-landing, and reactive-landing in collisions of protonated peptide ions with organic surfaces. Chem. Soc. Rev. 45, 3595–3608 (2016)
Hodgkin, D.C., Oughton, B.M.: Possible molecular models for gramicidin S and their relationship to present ideas of protein structure. Biochem. J. 65, 752–756 (1957)
Kupser, P., Pagel, K., Oomens, J., Polfer, N., Koksch, B., Meijer, G., von Helden, G.: Amide-I and -II vibrations of the cyclic β-sheet model peptide gramicidin S in the gas phase. J. Am. Chem. Soc. 132, 2085–2093 (2010)
Cai, S., Singh, B.R.: Identification of B-turn and random coil amide III infrared bands for secondary structure estimation of proteins. Biophys. Chem. 80, 7–20 (1999)
Cai, S., Singh, B.R.: A distinct utility of the amide iii infrared band for secondary structure estimation of aqueous protein solutions using partial least squares methods. Biochemistry 43, 2541–2549 (2004)
Myrskog, A., Anderson, H., Aastrup, T., Ingemarsson, B., Liedberg, B.: Esterification of self-assembled carboxylic-acid-terminated thiol monolayers in acid environment: a time-dependent study. Langmuir 26, 821–829 (2010)
Arnold, R., Azzam, W., Terfort, A., Wöll, C.: Preparation, modification, and crystallinity of aliphatic and aromatic carboxylic acid terminated self-assembled monolayers. Langmuir 18, 3980–3992 (2002)
Fallah, M.A., Stanglmair, C., Pacholski, C., Hauser, K.: Devising self-assembled-monolayers for surface-enhanced infrared spectroscopy of pH-driven poly-L-lysine conformational changes. Langmuir 32, 7356–7364 (2016)
Acknowledgements
This work was supported by the U.S. Department of Energy (DOE), Office of Basic Energy Sciences, Chemical Sciences, Geosciences and Biosciences Division. The research was performed using EMSL, a national scientific user facility sponsored by the DOE’s Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory (PNNL). PNNL is operated by Battelle for DOE under Contract DE-AC05-76RL01830.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 337 kb)
Rights and permissions
About this article
Cite this article
Laskin, J., Hu, Q. Reactive Landing of Gramicidin S and Ubiquitin Ions onto Activated Self-Assembled Monolayer Surfaces. J. Am. Soc. Mass Spectrom. 28, 1304–1312 (2017). https://doi.org/10.1007/s13361-017-1614-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-017-1614-2