Abstract
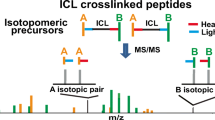
Chemical cross-linking of proteins followed by proteolysis and mass spectrometric analysis of the resulting cross-linked peptides provides powerful insight into the quaternary structure of protein complexes. Mixed-isotope cross-linking (a method for distinguishing intermolecular cross-links) was coupled with liquid chromatography, ion mobility spectrometry and mass spectrometry (LC-IMS-MS) to provide an additional separation dimension to the traditional cross-linking approach. This method produced multiplet m/z peaks that are aligned in the IMS drift time dimension and serve as signatures of intermolecular cross-linked peptides. We developed an informatics tool to use the amino acid sequence information inherent in the multiplet spacing for accurate identification of the cross-linked peptides. Because of the separation of cross-linked and non-cross-linked peptides in drift time, our LC-IMS-MS approach was able to confidently detect more intermolecular cross-linked peptides than LC-MS alone.




Similar content being viewed by others
References
Rappsilber, J.: The beginning of a beautiful friendship: Cross-linking/mass spectrometry and modelling of proteins and multi-protein complexes. J. Struct. Biol. 173(3), 530–540 (2011)
Sinz, A.: Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein-protein interactions. Mass Spectrom. Rev. 25(4), 663–682 (2006)
Leitner, A., Walzthoeni, T., Kahraman, A., Herzog, F., Rinner, O., Beck, M., Aebersold, R.: Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Mol. Cell. Proteomics. 9(8), 1634–1649 (2010)
Schilling, B., Row, R.H., Gibson, B.W., Guo, X., Young, M.M.: MS2Assign, automated assignment and nomenclature of tandem mass spectra of chemically crosslinked peptides. J. Am. Soc. Mass Spectrom. 14(8), 834–850 (2003)
Young, M.M., Tang, N., Hempel, J.C., Oshiro, C.M., Taylor, E.W., Kuntz, I.D., Gibson, B.W., Dollinger, G.: High throughput protein fold identification by using experimental constraints derived from intramolecular cross-links and mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 97(11), 5802–5806 (2000)
Walzthoeni, T., Claassen, M., Leitner, A., Herzog, F., Bohn, S., Forster, F., Beck, M., Aebersold, R.: False discovery rate estimation for cross-linked peptides identified by mass spectrometry. Nat. Methods 9(9), 901–903 (2012)
Yang, B., Wu, Y.-J., Zhu, M., Fan, S.-B., Lin, J., Zhang, K., Li, S., Chi, H., Li, Y.-X., Chen, H.-F., Luo, S.-K., Ding, Y.-H., Wang, L.-H., Hao, Z., Xiu, L.-Y., Chen, S., Ye, K., He, S.-M., Dong, M.-Q.: Identification of cross-linked peptides from complex samples. Nat. Methods 9(9), 904–906 (2012)
Herzog, F., Kahraman, A., Boehringer, D., Mak, R., Bracher, A., Walzthoeni, T., Leitner, A., Beck, M., Hartl, F.-U., Ban, N., Malmström, L., Aebersold, R.: Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science 337(6100), 1348–1352 (2012)
Taverner, T., Hall, N.E., O'Hair, R.A.J., Simpson, R.J.: Characterization of an antagonist interleukin-6 dimer by stable isotope labeling, cross-linking, and mass spectrometry. J. Biol. Chem. 277(48), 46487–46492 (2002)
Santos, L.F.A., Iglesias, A.H., Pilau, E.J., Gomes, A.F., Gozzo, F.C.: Traveling-Wave ion mobility mass spectrometry analysis of isomeric modified peptides arising from chemical cross-linking. J. Am. Soc. Mass Spectrom. 21(12), 2062–2069 (2010)
Smith, D.P., Anderson, J., Plante, J., Ashcroft, A.E., Radford, S.E., Wilson, A.J., Parker, M.J.: Trifluoromethyldiazirine: an effective photo-induced cross-linking probe for exploring amyloid formation. Chem. Commun. (44), 5728–5730 (2008)
Kindy, J.M., Taraszka, J.A., Regnier, F.E., Clemmer, D.E.: Quantifying peptides in isotopically labeled protease digests by ion mobility/time-of-flight mass spectrometry. Anal. Chem. 74(5), 950–958 (2002)
Soderberg, C.A.G., Lambert, W., Kjellstrom, S., Wiegandt, A., Wulff, R.P., Mansson, C., Rutsdottir, G., Emanuelsson, C.: Detection of Crosslinks within and between Proteins by LC-MALDI-TOFTOF and the Software FINDX to Reduce the MSMS-Data to Acquire for Validation. PLoS One 7(6), e38927 (2012)
Acton, T.B., Xiao, R., Anderson, S., Aramini, J., Buchwald, W.A., Ciccosanti, C., Conover, K., Everett, J., Hamilton, K., Huang, Y.J., Janjua, H., Kornhaber, G., Lau, J., Lee, D.Y., Liu, G.H., Maglaqui, M., Ma, L.C., Mao, L., Patel, D., Rossi, P., Sahdev, S., Shastry, R., Swapna, G.V.T., Tang, Y.F., Tong, S.C., Wang, D.Y., Wang, H., Zhao, L., Montelione, G.T.: Preparation of protein samples for NMR structure, function, and small-molecule screening studies. Methods Enzymol. 493, 21–60 (2011)
Cort, J.R., Selan, U., Schulte, A., Grimm, F., Kennedy, M.A., Dahl, C.: Allochromatium vinosum DsrC: solution-state NMR structure, redox properties, and interaction with DsrEFH, a protein essential for purple sulfur bacterial sulfur oxidation. J. Mol. Biol. 382(3), 692–707 (2008)
Livesay, E.A., Tang, K., Taylor, B.K., Buschbach, M.A., Hopkins, D.F., LaMarche, B.L., Zhao, R., Shen, Y., Orton, D.J., Moore, R.J., Kelly, R.T., Udseth, H.R., Smith, R.D.: Fully Automated four-column capillary LC-MS system for maximizing throughput in proteomic analyses. Anal. Chem. 80(1), 294–302 (2007)
Ibrahim, Y., Belov, M.E., Tolmachev, A.V., Prior, D.C., Smith, R.D.: Ion funnel trap interface for orthogonal time-of-flight mass spectrometry. Anal. Chem. 79(20), 7845–7852 (2007)
Ibrahim, Y.M., Prior, D.C., Baker, E.S., Smith, R.D., Belov, M.E.: Characterization of an ion mobility-multiplexed collision-induced dissociation-tandem time-of-flight mass spectrometry approach. Int. J. Mass spectrom. 293(1/3), 34–44 (2010)
Jaitly, N., Mayampurath, A., Littlefield, K., Adkins, J. N., Anderson, G. A., Smith, R. D.: Decon2LS: an open-source software package for automated processing and visualization of high resolution mass spectrometry data. BMC Bioinforma. 10(87), (2009)
Crowell K.L., Shah, A.R., Slysz, G., Lamarche, B.L., Meng, D., Baker, E.S., Monroe, M.E., Anderson, G.A., Smith, R.D.: LC-IMS-MS feature finder: detecting multidimensional features in IMS-TOF MS data. Proceedings of the American Society for Mass Spectrometry Conference, Denver, CO (June 2011).
Mädler, S., Bich, C., Touboul, D., Zenobi, R.: Chemical cross-linking with NHS esters: a systematic study on amino acid reactivities. J. Mass Spectrom. 44(5), 694–706 (2009)
Kalkhof, S., Sinz, A.: Chances and pitfalls of chemical cross-linking with amine-reactive N-hydroxysuccinimide esters. Anal. Bioanal. Chem. 392, 305–312 (2008)
Kahraman, A., Malmström, L., Aebersold, R.: Xwalk: computing and visualizing distances in cross-linking experiments. Bioinformatics 27(15), 2163–2164 (2011)
Leitner, A., Reischl, R., Walzthoeni, T., Herzog, F., Bohn, S., Förster, F., Aebersold, R.: Expanding the chemical cross-linking toolbox by the use of multiple proteases and enrichment by size exclusion chromatography. Mol. Cell. Proteom. 11(3) (2012)
Chen, Z.A., Jawhari, A., Fischer, L., Buchen, C., Tahir, S., Kamenski, T., Rasmussen, M., Lariviere, L., Bukowski-Wills, J.-C., Nilges, M., Cramer, P., Rappsilber, J.: Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. EMBO J. 29(4), 717–726 (2010)
Fabris, D., Yu, E.T.: Elucidating the higher-order structure of biopolymers by structural probing and mass spectrometry: MS3D. J. Mass Spectrom. 45(8), 841–860 (2010)
Li, H., Wells, S.A., Jimenez-Roldan, J.E., Römer, R.A., Zhao, Y., Sadler, P.J., O'Connor, P.B.: Protein flexibility is key to cisplatin crosslinking in calmodulin. Protein Sci. 21(9), 1269–1279 (2012)
Valentine, S.J., Counterman, A.E., Hoaglund, C.S., Reilly, J.P., Clemmer, D.E.: Gas-phase separations of protease digests. J. Am. Soc. Mass Spectrom. 9(11), 1213–1216 (1998)
Woods, A., Koomen, J., Ruotolo, B., Gillig, K., Russel, D., Fuhrer, K., Gonin, M., Egan, T., Schultz, J.: A study of peptide–peptide interactions using MALDI ion mobility o-TOF and ESI mass spectrometry. J. Am. Soc. Mass Spectrom. 13(2), 166–169 (2002)
Baker, E.S., Livesay, E.A., Orton, D.J., Moore, R.J., Danielson, W.F., Prior, D.C., Ibrahim, Y.M., LaMarche, B.L., Mayampurath, A.M., Schepmoes, A.A., Hopkins, D.F., Tang, K., Smith, R.D., Belov, M.E.: An LC-IMS-MS platform providing increased dynamic range for high-throughput proteomic studies. J. Proteome Res. 9(2), 997–1006 (2009)
Acknowledgments
The authors thank Dr. Gaetano Montelione and the NESG for providing the plasmid construct for expressing SrfN/STM0082 and the mixture of unlabeled and 13C,15N-labeled SO_2176, Dr. Sam Payne and Dr. Gordon Slysz for helpful discussions, and Dr. Abdullah Kahraman for assistance with the Xwalk program. This research was supported by the National Institute of General Medical Sciences (NIGMS grant GM094623). SrfN protein was produced in a project funded by the National Institute of Allergy and Infectious Diseases (IAA Y1-AI-8401-01). The work used instrumentation and capabilities developed with support from the NIGMS grant 8 P41 GM103493-10, and the US Department of Energy/Office of Biological and Environmental Research (DOE/BER). This work was performed in EMSL, a DOE/BER National Scientific User Facility located at PNNL in Richland, Washington.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 2423 kb)
Rights and permissions
About this article
Cite this article
Merkley, E.D., Baker, E.S., Crowell, K.L. et al. Mixed-Isotope Labeling with LC-IMS-MS for Characterization of Protein–Protein Interactions by Chemical Cross-Linking. J. Am. Soc. Mass Spectrom. 24, 444–449 (2013). https://doi.org/10.1007/s13361-012-0565-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-012-0565-x