Abstract
Significant differences in the two-dimensional electrophoresis patterns of proteins from developing rye grain were found to be associated with resistance and susceptibility to preharvest sprouting (PHS). Mass spectrometry of individual spots showing different abundance in PHS-resistant and PHS-susceptible lines identified proteins involved in: reaction to biotic and abiotic stresses, including oxidative stress, energy metabolism and regulation of gene expression. Highly differentiated abundance of proteins found in developing grain suggest that the diversification of processes leading to developing PHS resistance or PHS susceptibility starts from an early stage of grain development. A part of the identified proteins in rye grain were also reported to be associated with PHS in wheat and rice, which suggests that some mechanisms affecting precocious germination might be common for different cereal species.
Similar content being viewed by others
Introduction
Preharvest sprouting (PHS) is a major factor negatively affecting the quality of crops in years with high precipitation during a period of grain maturation. Consequently, selection for genotypes showing no precocious germination in conditions of water stress is one of the priorities in many breeding programmes. This goal is rather difficult to achieve because PHS is a complex trait largely dependent on weather conditions (intensity and length of rain periods, profiles of temperatures) and key genes for strong PHS resistance have not yet been cloned. Genomic studies carried out in major cereal species lead to the conclusion that PHS and dormancy have a complex genetic basis, with numerous quantitative trait loci (QTL) distributed across the majority of chromosomes (Gale et al. 2002). Recently, Rehman Arif et al. (2012) dissected PHS and dormancy in wheat into 18 chromosome bins, each containing a cluster of several genes.
PHS-related genes, even if they are clustered with other loci, may be identified by their different expression at the mRNA and/or protein level in PHS-resistant and PHS-susceptible grain, since these contrasting phenotypes should be determined by specific profiles of genes expression and protein activities during development. This hypothesis was partially proved in wheat (Bykova et al. 2011) and rye (Masojć and Kosmala 2012) by using two-dimensional gel electrophoresis (2-DE) and mass spectrometry (MS) to compare protein composition in mature grain of PHS-resistant and PHS-susceptible lines derived from a single cross. Both investigations revealed associations between PHS resistance and the abundance of specific proteins involved in responses to biotic and abiotic stresses, including oxidative stress. Oxidative stress caused by the accumulation of reactive oxygen species (ROS) in embryo cells was shown to be correlated with dormancy alleviation and germination (Oracz et al. 2009; Bykova et al. 2011). This may explain the different accumulation of ROS-scavenging enzymes in PHS-resistant and PHS-susceptible wheat (Bykova et al. 2011) and rye (Masojć and Kosmala 2012) grain.
Rye proteome (Masojć and Kosmala 2012) and genome (Masojć and Milczarski 2009; Masojć et al. 2009) studies show that PHS resistance has a complex genetic basis involving at least 24 proteins of various functions and numerous QTL distributed on each of the seven chromosomes. This knowledge should be enriched with proteomic studies carried out on different stages of grain development. It is expected that proteome analysis of developing grain will generate more information on molecular mechanisms affecting predisposition to PHS.
This investigation is aimed at finding differences in protein composition between PHS-resistant and PHS-susceptible lines of rye at the early stages of grain development.
Materials and methods
Two sets of rye recombinant inbred lines (RILs), 20 PHS-resistant lines (0 % sprouting in spikes after 7 days of water spray at maturity) and 20 PHS-susceptible lines (90–100 % sprouting in spikes after 7 days of water spray at maturity), were grown on the experimental field of the West Pomeranian University of Technology, Szczecin in 2011. These lines represented F10 generation of RILs developed from the 541 × Ot1-3 intercross described in earlier studies (Masojć et al. 2009; Masojć and Kosmala 2012). Two spikes per line were immersed in liquid nitrogen at the 25th day after anthesis (25 DPA) and kept at −30 °C for 1 month. Frozen kernels with green pericarp and milky endosperm (10 per line) were threshed out from spikes and freeze-dried in an Alpha 1-2 LD plus freeze drier (Christ®). Dry kernels of the PHS-resistant group of lines were bulked and ground in a Retsch MM 200 mill. Kernels from the PHS-susceptible group of lines were bulked and ground in the same mill. Two bulked samples of the rye flour representing 20 PHS-resistant and 20 PHS-susceptible lines were kept in a freezer prior to protein extraction.
The proteomic work involved: (1) the analyses of rye grain protein abundance in the bulked samples of the resistant and the susceptible rye inbred lines using 2-DE, (2) MS identification of proteins which were differentially accumulated between the analysed bulks. The protocol for proteomic research was the same as that described in detail by Masojć and Kosmala (2012).
Protein extraction was performed according to the method described by Hurkman and Tanaka (1986). This method was shown earlier to be useful to extract the proteins from different species and its use resulted in high-quality 2-D maps with proteins derived from different cell components, including membranes, chloroplasts, mitochondria and cytoplasm (Kosmala et al. 2009, 2012; Winiarczyk and Kosmala 2009; Bocian et al. 2011). Briefly, the powdered tissue was homogenised with 500 μl of extraction buffer (0.7 M sucrose, 0.5 M Tris, 30 mM HCl, 50 mM EDTA, 2 % DTT, 0.1 M KCl). An equal volume of water-saturated phenol was then added and the sample was incubated for 5 min at 4 °C, followed by 10 min of vortexing and centrifuging at 8,700 × g for 30 min. The phenol phase (upper) was recovered and re-extracted with an equal volume of extraction buffer. After centrifuging, the proteins from the phenol phase were precipitated by the addition of five volumes of cold 0.1 M ammonium acetate in methanol and then incubated at −20 °C at least overnight. The sample was then centrifuged at 20,500 × g at 0 °C for 30 min, the precipitate washed twice with the cold ammonium acetate in methanol and once in cold acetone, and dried. The pellet was finally dissolved in 100 μl of the sample solution (7 M urea, 2 M thiourea, 2 % NP-40, 2 % IPG buffer pH range 4–7 or 3–10, 40 mM DTT). The protein concentration was determined by the use of a 2-D Quant Kit (GE Healthcare). The aliquots of proteins extracted from 25 mg of rye flour were mixed with rehydration solution (7 M urea, 2 M thiourea, 2 % NP-40, 0.5 % IPG buffer pH range 4–7, 0.002 % bromophenol blue, 18 mM DTT) to the final volume of 450 μl and used for 2-DE, performed according to Hochstrasser et al. (1988). In the first dimension, isoelectric focusing (IEF), 24-cm Immobiline DryStrip gels with linear pH range 4–7 were used. This range was selected as the standard condition for resolving the proteins after the pre-selection performed with a relatively broad pH range (3–10) (data not shown) and followed our earlier work (Masojć and Kosmala 2012). Rehydration and focusing was carried out in Ettan IPGphor II (GE Healthcare) at 50 μA per strip at 20 °C, applying the following programme: 12 h of rehydration at 0 V and 9 h of focusing at 1 h/500 V, 2 h/1,000 V and 6 h/8,000 V. After IEF, the strips were equilibrated for 15 min in SDS equilibration buffer solution (6 M urea, 75 mM Tris–HCl pH = 8.8, 29.3 % glycerol, 2 % SDS, 0.002 % bromophenol blue, 65 mM DTT), followed for 15 min with the same buffer but containing 135 mM iodoacetamide instead of DTT. After equilibration, the proteins were separated in the second dimension, sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE), using 13 % polyacrylamide gels (1.5 × 255 × 196 mm) at 4 W/gel for 30 min and then at 17 W/gel for 4 h. Rainbow™ Molecular Weight Marker (GE Healthcare) was used as a standard to determine the molecular weights (MWs) of proteins for particular spots. Following electrophoresis, the gels were stained with colloidal Coomassie Brilliant Blue G-250, using the modified method of Neuhoff et al. (1988). Total separated protein spots on the gels were scanned by an ImageScanner III (GE Healthcare) and subjected to processing by the LabScan 6.0 program (GE Healthcare). Spot detection and image analyses (normalisation, spot matching, accumulation comparison) were performed with the ImageMaster 2D Platinum software package (GE Healthcare). To compensate for subtle differences in sample loading, gel staining and destaining, the abundance of each protein spot was normalised as a relative volume (% Vol). The % Vol of each spot was automatically calculated by the ImageMaster software as a ratio of the volume of a particular spot to the total volume of all the spots present on the gel. The extraction procedure and electrophoretic separation were performed twice (technical replicates), and the % Vol for the spots from the two replicated gels were then used to calculate the means, which were used to make comparisons between the analysed rye lines.
The protein spots which showed at least 2-fold (p < 0.05) differences in protein abundance between two analysed lines (quantitative analysis) together with protein spots present only in one of the analysed lines (qualitative analysis) were subjected to MS analysis and identification. Protein spots were excised from the gel and analysed by liquid chromatography coupled to the mass spectrometer in the Laboratory of Mass Spectrometry, Institute of Biochemistry and Biophysics, Polish Academy of Sciences (Warsaw, Poland). Samples were concentrated and desalted on an RP-C18 pre-column (Waters) and further peptide separation was achieved on a nano-Ultra Performance Liquid Chromatography (UPLC) RP-C18 column (Waters, BEH130 C18 column, 75 μm i.d., 250 mm long) of a nanoACQUITY UPLC system, using a 45-min linear acetonitrile gradient. The column outlet was directly coupled to the electrospray ionisation (ESI) ion source of an Orbitrap-type mass spectrometer (Thermo), working in the regime of data-dependent MS to MS/MS switch. An electrospray voltage of 1.5 kV was used. Raw data files were pre-processed with Mascot Distiller software (version 2.3, MatrixScience). The obtained peptide masses and fragmentation spectra were matched to the National Center Biotechnology Information (NCBI) non-redundant database with a Viridiplantae filter (1,032,142 sequences) using the Mascot search engine (Mascot Daemon v. 2.3, Mascot Server v. 2.2.03, MatrixScience). The following search parameters were applied: enzyme specificity was set to trypsin, peptide mass tolerance to ±40 ppm and fragment mass tolerance to ±0.8 Da. The protein mass was left as unrestricted and mass values as monoisotopic, with one missed cleavage being allowed. Alkylation of cysteine by carbamidomethylation as fixed and oxidation of methionine was set as a variable modification.
Protein identification was performed using the Mascot search probability-based MOWSE score. The ions score was −10* log(P), where P was the probability that the observed match was a random event. The Mascot-defined threshold, which indicated identity or extensive homology (p < 0.05), was 40 or less, therefore, an ion score of 40 was taken as a threshold for analysis. The proteins with the highest Multidimensional Protein Identification Technology (MudPIT) scores and/or the highest number of peptide sequences were selected. If more than one reliable identification appeared in the single spot, they were all indicated. However, in these cases, it was impossible to evaluate the abundance of particular proteins present in the spot and the total protein abundance was shown. Further research to estimate detailed protein contents of such multi-protein spots would be required according to, for example, the method described by Ishihama et al. (2005). When the protein was identified as “predicted protein”, its amino acid sequence was blasted using the blastp algorithm. The protein with the highest score was then selected as the functional homologue of the “predicted protein”.
Results
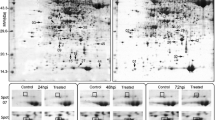
Following our earlier work (Masojć and Kosmala 2012), the pH = 4–7 range was selected as the standard condition for resolving the proteins to achieve the best compromise between the number and resolution of the separated spots. Only the spots which were detected within two replicate gels were included into the analyses. Out of all protein spots separated on 2-DE gels, 27 showed significant quantitative differences between PHS-resistant and PHS-susceptible lines (Figs. 1 and 2). Thirteen of them (spot nos. 2, 3, 4, 5, 6, 9, 10, 11, 14, 16, 18, 21, 22) were 2–4.1-fold more abundant in PHS-susceptible lines than in PHS-resistant lines. Fourteen others (spot nos. 1, 7, 8, 12, 13, 15, 17, 19, 20, 23, 24, 25, 26, 27) were 2–4.3-fold more abundant in PHS-resistant lines. Two protein spots (nos. 29 and 30) were found only in PHS-susceptible lines (Fig. 2), whereas spot no. 28 was detectable only in PHS-resistant lines (Fig. 1).
Two-dimensional gel electrophoresis (2-DE) map of proteins from developing rye grain [25th day after anthesis (25 DPA)] of 20 recombinant inbred lines (F10) highly resistant to pre-harvest sprouting (PHS), derived from the 541 × Ot1-3 cross. Proteins were separated by isoelectric focusing (IEF) at pH range 4–7, followed by polyacrylamide gel electrophoresis in sodium dodecyl sulphate (SDS-PAGE) and staining with colloidal Coomassie Brilliant Blue G-250. Spot detection and image analyses (normalisation, spot matching, protein accumulation analyses) were performed using the ImageMaster 2D Platinum software package (GE Healthcare). The normalised volumes of matched spots were used for comparisons made between the PHS-resistant and PHS-susceptible Secale cereale lines (in the case of each spot, the means derived from two replicated gels were applied). One protein spot (spot no. 28) present only in PHS-resistant lines and 27 protein spots (spot nos. 1–27) with quantitative differences in protein abundance between PHS-resistant and PHS-susceptible lines are indicated and numbered on the gel. The molecular weight (MW) and pH scales are shown
2-DE map of proteins from developing rye grain (25 DPA) of 20 recombinant inbred lines (F10) highly susceptible to PHS, derived from the 541 × Ot1-3 cross. Two protein spots (spot nos. 29 and 30) present only in PHS-susceptible lines and 27 protein spots (spot nos. 1–27) with quantitative differences in protein abundance between PHS-resistant and PHS-susceptible lines are indicated and numbered on the gel
MS of individual spots revealed that the majority of them were homogenous, containing one specific protein (Table 1). However, spot nos. 3, 4, 5, 6, 7, 9, 10, 11, 22, 25, 26, 27 and 28 were found to be heterogeneous, with 2 or 3 (in the case of spot no. 10) different proteins identified in each spot. In these spots, the relative abundance of particular proteins was not established. In total, 44 proteins were found in 30 spots, showing an association with PHS.
A predominant function of proteins more abundant in PHS-susceptible lines was defence against pathogens, as shown by serpin-Z1B (spot no. 6), cereal-type alpha-amylase inhibitor (spot no. 18), monomeric alpha-amylase inhibitor (spot nos. 21 and 22), basic endochitinase (spot no. 11) and vicilin-like antimicrobial peptides (spot nos. 9 and 10). Sensitivity to PHS was also associated with a higher amount of several enzymes that sustain energy metabolism (spot nos. 3, 4 and 5), resist abiotic stresses (spot nos. 6, 14 and 29) and act as ROS scavengers (spot nos. 2, 11 and 29).
Sprouting-resistant lines contained higher amounts of proteins protecting against: desiccation (spot no. 7), heat shock and other abiotic stresses (spot nos. 8, 13, 25 and 26) and pests (spot nos. 17, 20, 24, 27 and 28). Relatively higher abundance of dimeric alpha-amylase inhibitor (spot nos. 17 and 20), subtilisin-chymotrypsin inhibitor and triosephosphate isomerase (spot nos. 27 and 28), glycine-rich RNA-binding protein (spot no. 19) and xylanase inhibitor (spot no. 24) in developing grain was associated with resistance to PHS.
There were examples of the same proteins present in different spots, of which one molecular form was more abundant in PHS-resistant lines and another was associated with PHS-susceptible lines (16.9-kDa class I heat shock protein 1, spot nos. 14 and 15; peroxiredoxin, spot nos. 11 and 12; histone H4, spot nos. 26 and 30). On the other hand, multiple spots for one protein more abundant in PHS-resistant lines (dimeric alpha-amylase inhibitor, spot nos. 17 and 20; chaperonin GroEL, spot nos. 25 and 26; elongation factor Tu, spot nos. 25 and 27) or in PHS-susceptible lines (monomeric alpha-amylase inhibitor, spot nos. 21 and 22; vicilin-like antimicrobial peptides, spot nos. 9 and 10) were also found.
The majority of the detected proteins were involved in response to abiotic and biotic stresses and in energy metabolism, which suggest that predisposition to PHS is, above all, a genetically determined molecular mechanism of response to various kinds of stresses acting during grain desiccation followed by prolonged contact with water. Since the inability of mature grain to germinate in water-sprayed spikes showed by PHS-resistant lines should be imposed by the genetic control of down- and/or up-regulation of genes expression, it is not surprising that some regulatory proteins were revealed in this study. They were found in spot no. 7 (14-3-3 regulatory protein), spot no. 9 (ras-related protein RIC2-like), spot no. 19 (glycine-rich RNA-binding protein) and in spot nos. 25 and 27 (elongation factor Tu). Beta1 proteasome found in spot no. 10 is another example of a protein whose proteolytic activity may influence other proteins’ composition and activity within different parts of developing grain. The consequences for gene expression on the genomic scale might result from apparently substantial differences in the abundance of histone H4 isoforms, which control chromatin structure and DNA–protein interaction. One histone H4 isoform (spot no. 30) was found only in PHS-susceptible lines, whereas the second (spot no. 26) was shown to be more abundant in PHS-resistant lines.
At the present stage of knowledge on molecular mechanisms leading to PHS resistance, it is difficult to explain the different abundances of three other proteins in the grain of PHS-susceptible and PHS-resistant lines. A single molecular form of RuBisCO (spot no. 1), a basic enzyme for carbon fixation during photosynthesis taking place in the green pericarp of developing grain, showed a relatively high level of accumulation in the PHS-resistant group of lines. Similarly, early-methionine-labelled polypeptide of unknown function (spot no. 23) was found to be associated with high resistance to PHS. On the other hand, the developing grain of PHS-susceptible lines contained more aspartate-semialdehyde dehydrogenase (spot no. 4), an enzyme involved in lysine biosynthesis.
Discussion
Differences in protein composition of the grain at 25 DPA, found between the two groups of rye lines with extremely different predispositions to PHS, clearly show that the process of preparing grain to resist germination in mature spike during the rainy season starts in early development (this paper) and continues up to the end of grain maturation (Masojć and Kosmala 2012). Proteomes of PHS-resistant and PHS-susceptible grain revealed more differences in protein abundance 25 days post-anthesis than at the maturity stage. There were, however, several common features of the PHS-related protein patterns at both stages of grain development. In both studies, PHS-resistant grain contained higher levels of dimeric alpha-amylase inhibitor and PHS-susceptible grain had increased levels of ATP synthase, serpin-Z1B and monomeric alpha-amylase inhibitor. Differences in the abundance of 16.9-kDa class I heat-shock protein, triosephosphate isomerase and 14-3-3 regulatory protein were depicted in both studied stages of seed formation. Generally, both young and mature seeds of PHS-resistant and PHS-susceptible lines showed differentiation in respect to proteins responding to biotic and abiotic stresses, including oxidative stress. The set of differentially accumulated proteins related to biotic stress showed higher complexity in developing seeds. Similarly, a more complex set of proteins affecting genes expression and protein synthesis and degradation rates was associated with PHS at the early stage of grain development. This observation raises the question about the significance of transiently expressed differences in protein abundance during grain development for attaining PHS resistance or susceptibility at maturity. There are two possible answers: (1) differences observed at 25 DPA persist until seed maturity but they decrease below the 2-fold threshold value for 2-DE analysis and (2) differences in concentrations of particular proteins decline with the time but they have a long-lasting impact on the grain properties.
Similarly to in a previous study (Masojć and Kosmala 2012), individual proteins associated with PHS were detected in different spots separated by means of 2-DE. Various reasons might underlie the multiple molecular forms of proteins, including multi-gene families (isoforms), heterozygosity (allozymes), post-translational modifications or even protein degradation during sample preparation. Change in protein abundance due to different frequencies of allelic variants within each of the two extreme groups of lines would be an indirect proof for the involvement of respective structural genes in PHS resistance/susceptibility. Verification of allelic forms among 2-DE protein spots should be possible by comparing patterns of individual RILs, including parental lines.
It is known that resistance to PHS can be reduced when seed coat is ruptured by pests at any stage of development. The persistence of dimeric alpha-amylase inhibitor in higher amounts in developing and mature grain of PHS-resistant lines and higher amounts of xylanase inhibitor and subtilisin-chymotrypsin inhibitor at 25 DPA gives strong evidence that the accumulation of these particular defence proteins is important for the PHS resistance of rye. Different sets of defence proteins seem to be important for PHS susceptibility, since it is associated with a higher abundance of serpins and monomeric alpha-amylase inhibitor at both 25 DPA and maturity, and with increased levels of immunophilin and endochitinase C at 25 DPA and tritin at maturity.
Another specific feature of PHS-susceptible lines detected in both proteomic studies of rye grain is the relatively higher accumulation level of some proteins related to energy metabolism, including those involved in the citric acid cycle and in glycolysis. This is, above all, ATP synthase overproduced at 25 DPA and at maturity, glucose and ribitol dehydrogenase, triosephosphate isomerase and phosphoglucomutase (maturity), phosphoglycerate kinase, succinyl-CoA ligase, fructose–bisphosphate aldolase (at 25 DPA). Elevated synthesis of these proteins might be necessary to achieve unusually high rates of germination observed in PHS-susceptible RILs. A developmental programme of predisposing grain to PHS can also be based on different rates of protein degradation due to the activity of proteasomes. This hypothesis is supported by the detection of beta1 proteasome 7D protein among those which are up-regulated in PHS-susceptible lines at 25 DPA.
A higher level of anti-oxidant protection is seemingly attributed to the grain of PHS-susceptible lines, which accumulated ROS-scavenging enzymes, such as: glutathione transferase, dehydroascorbate reductase and superoxide dismutase (mature grain), but also produced possibly more L-ascorbic acid due to the up-regulation of UTP-glucose-1-phosphate uridylyltransferase synthesis at 25 DPA. Seed imbibition in water followed by the germination process is connected with the production of high levels of ROS (Bykova et al. 2011; Oracz et al. 2009), which might negatively affect young tissues of the seedling. It is possible that increased levels of ROS-scavenging enzymes in PHS-susceptible grain play a protective role during premature germination.
Many proteins protecting seeds against desiccation, low temperature and heat shock, including chaperones for other proteins, were found to be differentially distributed in PHS-resistant and PHS-susceptible grain at the 25 DPA. Among them, one isoform of 16.9-kDa heat shock protein (spot no. 14) was overproduced in PHS-susceptible grain and another isoform (spot no. 15) in PHS-resistant grain. This protein also showed higher abundance in PHS-susceptible mature grain (Masojć and Kosmala 2012). Evidently, rye lines extremely different in respect to PHS exhibited individual profiles of anti-stress protein synthesis during grain development. It is especially interesting in respect to proteins acting against desiccation stress, since PHS-resistant lines must be prepared to pass more rounds of seed imbibitions and desiccation before their dormancy is alleviated. Therefore, LEA protein (spot no. 7), HSP70 (spot. 8), GroEL chaperone (spot nos. 25 and 26) and SORBIDRAFT protein (spot no. 13) should be further evaluated in respect to their role in sprouting resistance.
More examples of regulatory proteins being differentially accumulated in the grain of PHS-resistant and PHS-susceptible lines were revealed through proteomic analysis performed at 25 DPA in respect to that carried out on mature grain (Masojć and Kosmala 2012). Identified proteins can affect other genes expression at different levels, such as transcription (14-3-3, ras-related protein RIC2-like), post-transcription (glycine-rich RNA-binding protein), translation (elongation factor Tu), post-translation (beta1 proteasome) and epigenetically (histone H4). The different molecular compositions of these potent proteins at early stages of grain development could have a strong impact on later molecular processes, leading to extreme phenotypes in respect to PHS.
Although wheat and rye could have evolved quite different strategies of PHS resistance, it is interesting to compare proteomic (Bykova et al. 2011) and genomic (Rehman Arif et al. 2012) analyses of this trait performed separately for each of these related species. As it was shown in Table 1, there are several genes and proteins which are common for wheat and rye as candidate genes for PHS resistance/susceptibility. This list contains serpins, heat shock protein HSP70, endochitinase, alpha-amylase inhibitors, subtilisin-chymotrypsin inhibitors, vicilin-like antimicrobial peptides, i.e. a complex set of proteins involved in plant response to biotic and abiotic stresses. Proteins found to be related to PHS in both wheat and rye are also peroxiredoxin, xylanase inhibitor and RNA-binding protein (Kamal et al. 2009, and this paper). In addition, a study on transcriptomes from rice grain representing cultivars of high and low dormancy (Qin et al. 2010) showed differences in the expression level of genes encoding heat shock proteins, which were also associated with PHS in wheat (Bykova et al. 2011) and rye (Masojć and Kosmala 2012, and this paper). Further work on genes controlling PHS should combine a map-based approach with the analysis of seed proteome and/or transcriptome. These potent research tools will lead to the identification of genes, their expression profiles and interactions, which are necessary in order to understand the molecular biology of PHS.
References
Bocian A, Kosmala A, Rapacz M, Jurczyk B, Marczak Ł, Zwierzykowski Z (2011) Differences in leaf proteome response to cold acclimation between Lolium perenne plants with distinct levels of frost tolerance. J Plant Physiol 168:1271–1279
Bykova NV, Hoehn B, Rampitsch C, Hu J, Stebbing J-A, Knox R (2011) Thiol redox-sensitive seed proteome in dormant and non-dormant hybrid genotypes of wheat. Phytochemistry 72:1162–1172
Gale MD, Flintham JE, Devos KM (2002) Cereal comparative genetics and preharvest sprouting. Euphytica 126:21–25
Hochstrasser DF, Harrington M, Hochstrasser AC, Miller MJ, Merril CR (1988) Methods for increasing the resolution of two-dimensional protein electrophoresis. Anal Biochem 173:424–435
Hurkman WJ, Tanaka CK (1986) Solubilization of plant membrane proteins for analysis by two-dimensional gel electrophoresis. Plant Physiol 81:802–806
Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M (2005) Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics 4:1265–1272
Kamal AHM, Kim KH, Shin DH, Seo HS, Shin KH, Park GS, Heo HY, Woo SH (2009) Proteomics profile of pre-harvest sprouting wheat by using MALDI-TOF mass spectrometry. Plant Omics 2:110–119
Kosmala A, Bocian A, Rapacz M, Jurczyk B, Zwierzykowski Z (2009) Identification of leaf proteins differentially accumulated during cold acclimation between Festuca pratensis plants with distinct levels of frost tolerance. J Exp Bot 60:3595–3609
Kosmala A, Perlikowski D, Pawłowicz I, Rapacz M (2012) Changes in the chloroplast proteome following water deficit and subsequent watering in a high- and a low-drought-tolerant genotype of Festuca arundinacea. J Exp Bot 63:6161–6172
Masojć P, Kosmala A (2012) Proteomic analysis of preharvest sprouting in rye using two-dimensional electrophoresis and mass spectrometry. Mol Breed 30:1355–1361
Masojć P, Milczarski P (2009) Relationship between QTLs for preharvest sprouting and alpha-amylase activity in rye grain. Mol Breed 23:75–84
Masojć P, Lebiecka K, Milczarski P, Wiśniewska M, Łań A, Owsianicki R (2009) Three classes of loci controlling preharvest sprouting in rye (Secale cereale L.) discerned by means of bidirectional selective genotyping (BSG). Euphytica 170:123–129
Neuhoff V, Arold N, Taube D, Ehrhardt W (1988) Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 9:255–262
Oracz K, El-Maarouf-Bouteau H, Kranner I, Bogatek R, Corbineau F, Bailly C (2009) The mechanisms involved in seed dormancy alleviation by hydrogen cyanide unravel the role of reactive oxygen species as key factors of cellular signaling during germination. Plant Physiol 150:494–505
Qin H, Wu F, Xie K, Cheng Z, Guo X, Zhang X, Wang J, Lei C, Wang J, Mao L, Jiang L, Wan J (2010) Transcriptomics analysis identified candidate genes colocalized with seed dormancy QTLs in rice (Oryza sativa L.). J Plant Biol 53:330–337
Rehman Arif MA, Neumann K, Nagel M, Kobiljski B, Lohwasser U, Börner A (2012) An association mapping analysis of dormancy and pre-harvest sprouting in wheat. Euphytica 188:409–417. doi:10.1007/s10681-012-0705-1
Winiarczyk K, Kosmala A (2009) Development of the female gametophyte in the sterile ecotype of the bolting Allium sativum L. Sci Hortic 121:353–360
Acknowledgements
This study was supported by the National Centre of Science in Poland, grant no. N N302 016139.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 102 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Masojć, P., Kosmala, A. & Perlikowski, D. Proteomic analysis of developing rye grain with contrasting resistance to preharvest sprouting. J Appl Genetics 54, 11–19 (2013). https://doi.org/10.1007/s13353-012-0127-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-012-0127-8