Abstract
Cornelia de Lange syndrome (CdLS) is a rare multi-system genetic disorder characterised by growth and developmental delay, distinctive facial dysmorphism, limb malformations and multiple organ defects. The disease is caused by mutations in genes responsible for the formation and regulation of cohesin complex. About half of the cases result from mutations in the NIPBL gene coding delangin, a protein regulating the initialisation of cohesion. To date, approximately 250 point mutations have been identified in more than 300 CdLS patients worldwide. In the present study, conducted on a group of 64 unrelated Polish CdLS patients, 25 various NIPBL sequence variants, including 22 novel point mutations, were detected. Additionally, large genomic deletions on chromosome 5p13 encompassing the NIPBL gene locus were detected in two patients with the most severe CdLS phenotype. Taken together, 42 % of patients were found to have a deleterious alteration affecting the NIPBL gene, by and large private ones (89 %). The review of the types of mutations found so far in Polish patients, their frequency and correlation with the severity of the observed phenotype shows that Polish CdLS cases do not significantly differ from other populations.
Similar content being viewed by others
Introduction
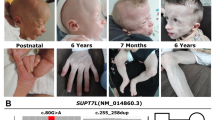
Cornelia de Lange syndrome (CdLS; OMIM#122470) is a rare autosomal dominant multi-system disorder with an estimated incidence of 1:10,000–1:50,000 live births. Although most of the cases occur sporadically, familial incidence has also been reported (Beck 1976, Russell et al. 2001; Barisic et al. 2008). The phenotype of patients with CdLS is very characteristic. The major clinical manifestations include a distinctive facial appearance (synophrys and high arched eyebrows in particular), prenatal and postnatal growth retardation, malformations of the extremities, especially the upper limbs, hirsutism, psychomotor retardation and a wide range of gastrointestinal problems (Gillis et al. 2004; Kline et al. 2007; Selicorni et al. 2007; Oliver et al. 2010). The severity of each of the aforesaid features differs significantly between the patients, which has prompted to define two CdLS subtypes: a mild form and the classical severe form (Kline et al. 2007; Oliver et al. 2010).
The disorder is referred to as a cohesinopathy, since it is caused by alterations in genes involved in the proper interaction of the cohesin complex, such as NIPBL, PDS5 (coding cohesion regulators) and SMC1, SMC3, RAD21 (coding proteins of the core of the cohesin complex) (Krantz et al. 2004; Tonkin et al. 2004; Zhang et al. 2009; Musio et al. 2006; Deardorff et al. 2007; Deardorff et al. 2012a). The primary function of the cohesin complex is to regulate chromosome segregation during cell division; however, cohesion is also implicated in the regulation of gene expression and DNA repair (Dorsett and Krantz 2009; Feeney et al. 2010). Accordingly, deleterious alteration of any of the genes coding proteins of the cohesin complex may affect early stages of prenatal development (Dorsett 2011).
The most common dysfunction of cohesin complex observed in CdLS patients is caused by mutations in the NIPBL gene, which is located on the chromosome 5p13.2 and consists of 47 exons. Its protein product is delangin, a 2804 aa human orthologue of fungal Scc2 and the Drosophila Nipped-B proteins. Delangin is involved in sister chromatid cohesion by interacting with the cohesin complex. The regulation of cohesin complex attachment to chromatids gives the possibility to control the expression of a number of genes (Liu et al. 2009; Bose and Gerton 2010). Homozygous mutations in the NIPBL gene had never been previously reported, providing further evidence of its crucial function in embryogenesis. Approximately 50 % of CdLS patients have a pathogenic mutation in the NIPBL gene, although the mutation detection rate differs considerably between various reports, ranging from 26 to 70 % (Borck et al. 2004; Gillis et al. 2004; Miyake et al. 2005; Yan et al. 2006; Bhuiyan et al. 2006; Schoumans et al. 2007; Selicorni et al. 2007; Kline et al. 2007; Pié et al. 2010).
To date, approximately 250 different point mutations have been identified in more than 300 CdLS patients (Leiden Open Variation Database; LODV: http://grenada.lumc.nl/LOVD2/CDLS/home.php?select_db=NIPBL). Alterations are localised along the entire coding sequence of the gene and, although some of them were reported in a few unrelated patients, no obvious hot spot has been found so far (Oliveira et al. 2010). A correlation between the type of point mutations and the severity of the phenotype has been observed. Truncating mutations are most commonly identified among patients with classical CdLS subtype, while missense mutations or small in-frame deletions/duplications more likely cause milder phenotype (Gillis et al. 2004; Yan et al. 2006; Pié et al. 2010; Oliveira et al. 2010). Moreover, in a few cases, a large genomic rearrangement involving the NIPBL gene has been detected, indicating even higher complexity of the genetic background of the syndrome. Observed genomic alterations were mostly deletions, encompassing one or more of NIPBL exons; however, also, a few pathogenic duplications were reported (Bhuiyan et al. 2007; Ratajska et al. 2010; Oliver et al. 2010; Murray et al. 2012; Pehlivan et al. 2012, Russo et al. 2012).
The aim of this study was to determine the prevalence and spectrum of the NIPBL gene point mutations among Polish CdLS patients diagnosed in the period 2006–2011. More to the point, patients negative for point mutations were subjected to further evaluation aimed at the identification of large genomic rearrangements (deletion/duplication of exons) within the NIPBL gene. The additional objective of the study was to compare the spectrum of mutations identified so far in Polish patients (Yan et al. 2006; Ratajska et al. 2010; Wierzba et al. 2011 and current results) with mutations described in CdLS patients from other populations.
Materials and methods
Sixty-four unrelated patients (34 females and 30 males) with clinical diagnosis of CdLS referred for genetic screening of the NIPBL gene between 2006 and 2011 through the Polish Cornelia de Lange Association were evaluated in the study. In accordance with the diagnostic criteria for CdLS (Kline et al. 2007), 31 (48 %) patients were classified as having classical (severe) phenotype and 33 (52 %) as having the mild form of CdLS.
DNA and, in selected cases, also RNA, was extracted from peripheral blood samples (Genomic Midi AX kit; Total RNA Mini kit; A&A Biotechnology). The complete coding sequence of the NIPBL gene, including splice junctions, was evaluated by a combination of the denaturing high-performance liquid chromatography (DHPLC) technique (WAVE DNA Fragment Analysis System, Transgenomic Inc.) and bi-directional sequencing (Applied Biosystems).
All novel variants were verified in the dbSNP, the 1000 Genomes catalogue and the NHLBI Exome Sequencing Project. Whenever available, de novo origin of the mutations was verified through parents’ testing. Besides, selected bioinformatics resources, including UCSC Genome Bioinformatics and UniProtKB tools, along with PolyPhen-2 and SIFT web interfaces, were used to assess the effect of the detected novel sequence variants on the structure and function of the protein. Potential effects on the splicing process were explored both in silico (Human Splicing Finder and ESEfinder web interfaces) and by reverse transcription polymerase chain reaction (RT-PCR) studies (Bioscript; Bioline Inc.).
Thirty-nine (39, 61 %) CdLS patients found to be negative for NIPBL point mutations were subjected to extended screening for genomic rearrangements using a Multiplex Ligation-dependent Probe Amplification (MLPA) and array-comparative genomic hybridization (aCGH) approach. PCR fragments from MLPA reactions performed using commercial kits P141 and P142 (MRC Holland) were analysed on an ABI PRISM 310 Genetic Analyzer (Applied Biosystems) using TAMRA-500 (Applied Biosystems) as the size standard. Raw data of each individual exon peak area were transferred to an Excel spreadsheet (Microsoft). Probe ratios below 0.7 were regarded as exons deletion and probe ratios above 1.3 as duplication. All abnormal results were repeated in a second independent experiment and, where confirmed, aCGH analysis of the given sample was performed using the Human Genome CGH Microarray 105A kit (Agilent Technologies). As the reference, DNA isolated from peripheral blood leukocytes of nine anonymous healthy females was used. The arrays were scanned with the Axon GenePix 4000B scanner and GenePix Pro 6.0 software (Molecular Devices); the results were analysed using Feature Extraction 9.5.3 software (Agilent Technologies). All procedures were performed according to the manufacturers’ instructions. More information on screening protocols, including the choice of primers, can be obtained from the corresponding author upon request.
Written informed consent was obtained from the relevant guardians of the children and from patients themselves, whenever eligible. The study was approved by the Ethical Committee of the Medical University of Gdansk, Poland (NKEBN/380/2006).
Results
Twenty-two (22, 34 %) patients were found to have novel private NIPBL sequence variants located along the entire coding sequence of the gene (Table 1). Besides, three previously described point mutations were detected in the analysed cohort of CdLS patients. The most common were non-synonymous sequence variants (n = 15), followed by deletions (n = 7), splice-site intronic mutations (n = 2) and a duplication (n = 1).
The 15 identified missense sequence variants, including two previously described mutations, were located along the entire coding sequence. The two known mutations were c.6892C > T and c.4321G > T (Table 1). In order to assess the pathogenic significance of the detected novel non-synonymous variants, in-silico structure–function analyses were attempted in all cases. BLAST search of the Protein Data Bank (http://www.pdb.org, 20.09.2012) with delangin sequence revealed no solved structures of proteins with considerable sequence similarity, except for the HEAT and PxVxL motifs. Seven substitutions affected evolutionarily conserved amino acid residues; however, none of them lay within five HEAT repeat domains, the crucial and highly conserved C-terminal portion of the protein.
Truncating mutations were the second most common type of mutations (n = 8). This subgroup included seven deletions and one duplication, all leading to reading frame shift, resulting in premature protein truncation. It should be underlined that all truncating mutations occurred in patients with classic type of the disease.
Three splice-site mutations, including the above presented exonic missense c.4321G > T and two novel intronic variants, were also detected. To confirm the pathogenic character of the novel variants, in-silico studies were performed. Moreover, in the case of c.3855 + 1delG, biological samples for RNA studies were also available. cDNA analysis confirmed that c.3855 + 1delG leads to exon 16 skipping. Even though there was no possibility to obtain blood samples for RNA analysis from the patient with the c.5863-1G > C variant, the position of this alteration strongly suggests its impact on the splicing process and a highly probable deletion of exon 33, as predicted by in-silico tools.
Patients with no NIPBL sequence alteration detected on direct sequencing were eligible for extended screening. No segmental indels within the gene were detected, while in two cases, MLPA analysis showed reduction of probe ratios below 0.6 for all exons, suggesting deletion of the entire NIPBL gene. The exact size and type of genomic alteration was further evaluated by aCGH. Both patients were found to have sub-microscopic interstitial deletions on the short arm of chromosome 5. The larger of the two deletions, of approximately 1.7 Mb, encompassed not only the entire NIPBL sequence, but also five other genes (namely, SLC1A3, FLJ1323, NUP155, WDR70, GDNF). The second patient was found to carry a deletion of about 0.65 Mb in size. In this case, a deleted region included NIPBL, FLJ1323, NUP155 and a part of the WDR70 gene.
Discussion
NIPBL gene mutations are the most common cause of CdLS worldwide, estimated to account for ca. 50 % of cases (Oliveira et al. 2010). The remaining patients have alterations in SMC1A (ca. 5 %), SMC3 (<1 %) and the recently discovered RAD21 and HDAC8 (Deardorff et al. 2007, 2012a, b). So far, the prevalence of mutations in the RAD21 and HDAC8 genes is rather anecdotal and large population studies are needed in order to evaluate their significance in the clinical setting (Deardorff et al. 2012a, b). Then again, a number of candidate genes (STAG2, ESCO1, ESCO2, PDS5 and MAU2) are subject to intensive studies aimed at proving their pathogenicity in humans, but no clinically relevant reports have been published so far.
Against this background, it should be underlined, however, that, despite the fact that several genes are indicated to be involved in CdLS, with NIPBL playing first fiddle, so far, in more than half of the patients, no genetic cause has been identified. At the current state of knowledge, while waiting for next-generation sequencing techniques to enter clinical practice, the optimal diagnostic screening strategy should include, in the first place, testing of the entire coding sequence of NIPBL, and only later referring the patient to more extended screening in line with the severity of the presented phenotype.
In the current study, the mutation detection yield of 42 % was similar to the mean value observed in the previously published reports, although the mutation detection rate differs considerably between published studies, ranging from 26 to 70 % (Borck et al. 2004; Gillis et al. 2004; Miyake et al. 2005; Yan et al. 2006; Bhuiyan et al. 2006; Schoumans et al. 2007; Selicorni et al. 2007; Kline et al. 2007; Pié et al. 2010). The observed discrepancies between the studies could result from the size of the group, accuracy of clinical diagnosis, selection of patients for the study or even differences in methods used for molecular analysis. Nevertheless, taking together all the results obtained so far from NIPBL testing in Polish patients (Table 1), the mutation detection rate remains relatively high (45 %) in the Polish population, providing further evidence of the clinical significance of NIPBL testing in all CdLS patients. Furthermore, we have found that 61 % of patients with severe phenotype and 24 % of children with mild form of the disease were positive for a mutation in the NIPBL gene. Taken together all the results obtained so far in the Polish cohort of patients, 64 % of classic CdLS cases and merely 20 % of mild ones had deleterious NIPBL alteration. Our findings are in line with the previously published reports, which point out the severity of NIPBL-associated CdLS phenotype, as shown by much higher mutation detection rates for this gene observed among severely affected children. More to the point, large genomic rearrangements encompassing the NIPBL locus were previously reported only in patients with classic form of the disease. This is also the case with respect to the current study, and to the Polish cohort in general, as we have detected interstitial deletions at chromosome 5p13.2 in 3 % of patients, all with the utmost severe phenotype.
Mutational analysis of the entire coding sequence of the NIPBL gene in the Polish cohort, both analyzed in the current study and analyzed together with the previous reports, confirmed that there is no evident hot spot. All identified mutations were private, except for seven (Table 1). For instance, our study is the first to report mutations in exon 24 of the gene. Our findings prove to be consistent with previous studies, as, so far, the most common mutation in CdLS patients, namely, p.Arg827Glyfs*2, was reported in seven (2 %) patients only (Oliveira et al. 2010). With regard to the seven recurrent mutations that were present both in a Polish patient and in previously reported patients from other ethnic and/or geographical origin, only in four (c.3060_3063delAGAG; c.4321G > T; c.5167C > T; c.7439_7440delGA) was the clinical picture of the disease similar to the phenotype presented by our patients (Gillis et al. 2004; Bhuiyan et al. 2006; Pié et al. 2010). Conversely, c.737A > G, c.6892C > T and c.6653_6655delATA have been identified in patients with varying phenotypes (Gillis et al. 2004; Borck et al. 2004; Selicorni et al. 2007; Kline et al. 2007). This suggests that, in the case of CdLS, the same genetic change does not always lead to the same degree of disease severity, a phenomenon which is probably influenced by additional, not yet specified, modifying factors (Gillis et al. 2004; Pié et al. 2010).
Over 250 various NIPBL point mutations have already been described in CdLS patients, the most common being missense mutations (28 %) and deletions leading to frame shift (23 %), followed by nonsense and split-site mutations (each 18 %), while less frequent variants are insertions and complex indels (Oliveira et al. 2010). Mutations detected in Polish patients follow a similar pattern with respect to the type and frequency of each of the categories. More to the point, the analysis of the subgroups of the patients distinguished with respect to the severity of the phenotype into classic and mild shows that truncating mutations and large genomic rearrangements occurred exclusively in the severe cases, while missense and splice-site mutations were observed in both groups of the patients. These results are in accordance with the literature data (Tonkin et al. 2004; Gillis et al. 2004; Schoumans et al. 2007; Pié et al. 2010; Oliveira et al. 2010).
In order to assess the pathogenic effect of the detected novel missense variants, a number of bioinformatical analyses has been performed. Instead of experimental verification of the potential presence of the detected variants in the control group of healthy volunteers, we have decided to check their incidence in two large exome sequence variant databases (1000 Genomes, NHLBI). Only one of the alterations present in our CdLS patients, namely, c.2603G > A, was found among benign variants (rs149629686) reported in these databases, with the frequency of the minor allele (MAF) being 0.04 %. Besides, c.535G > T affected the same nucleotide as a rare polymorphism c.535G > A reported in databases (rs142923613; MAF 0.2 %) but resulted in a different residue substitution (serine and not threonine). Six out of eight variants detected in the CdLS patients presenting severe classic phenotype were equally classified by SIFT and PolyPhen-2 algorithms to be deleterious and were found to affect evolutionary conserved residues. The two exceptions were c.2603G > A and c.5164A > C alterations. C.2603G > A was eventually called a mild variant, because not only was it found to be present in a 6/23,761 healthy individuals collected in the analysed exome databases, but also it affects a non-conserved residue in a non-structuralised fragment of the protein. Likewise, c.5164A > C affects a non-conserved residue and the analysis of the alignment reveals that at least five distinct amino acids with different physical characteristics occur at this site in various species. Nevertheless, both were de novo mutations, hence, we cannot explicitly decide on their pathogenic effect. Then again, five novel missense variants were reported in patients with mild form of CdLS. All but c.4873G > T, which affects a evolutionary conserved aminoacid residue, were classified as benign by the online in-silico tools; however, as no precise structural evaluation, due to the lack of solved structures of proteins with considerable sequence similarity, could be performed, it cannot be excluded that their presence is responsible for the mild phenotype observed in the patients. In the case of c.4909A > C, the replaced amino acid has similar physical characteristics, thus, the probability of its influence on the protein structure is rather low. Accordingly, these variants were categorized as mild, even though they were confirmed to be de novo.
Although CdLS is a monogenic disease, there have been over 30 cases of chromosomal abnormalities associated with that syndrome (DeScipio et al. 2005). In fact, a de novo balanced translocation involving chromosome 5 facilitated identification of the NIPBL gene (Tonkin et al. 2004). Recently, a few reports have been published presenting several cases with sub-microscopic deletions encompassing the NIPBL gene, with the size of aberration ranging from 4.2 kb (a single exon) to 2 Mb deletion, including not only the NIPBL gene but also 14 adjacent genes (Bhuiyan et al. 2007; Ratajska et al. 2010; Oliver et al. 2010, Murray et al. 2012; Pehlivan et al. 2012; Russo et al. 2012). The incidence of genomic aberrations was estimated to be 4–5 % of CdLS patients negative for the NIPBL point mutation (Pehlivan et al. 2012; Russo et al. 2012). Likewise, in the current study, we have detected two genomic alterations in the group of 39 Polish patients negative for point mutations in NIPBL (i.e. 5 %). Yet, the detection rate for the entire cohort of patients was <3 %, showing that, by and large, NIPBL alterations present in CdLS patients are point mutations. Both deletions occurred in patients with the most severe phenotype; however, no significant clinical feature atypical for CdLS was observed in these children.
Conclusions
In the current study, we have performed a comprehensive analysis of NIPBL alterations in a Polish cohort of Cornelia de Lange syndrome (CdLS) patients. Similarly to other populations and ethnic groups, no founder effect or hot spot within the gene were observed, as the prevailing type of mutations were novel private ones. In addition, we have confirmed the presence of large genomic rearrangements in the portion of the most severely affected patients; however, the detection rate of ∼5 % disqualifies the test from wide usage in clinical settings. We anticipate however, that the introduction of new diagnostic tools, next-generation sequencing in particular, which is at the point of entering clinical practice, will soon allow testing of all the postulated candidate genes involved in proper cohesion complex activity in the to-date undiagnosed portion of patients with respect to point mutations and to gene(s) dosage.
References
Barisic I, Tokic V, Loane M, Bianchi F, Calzolari E, Garne E, Wellesley D, Dolk H; EUROCAT Working Group (2008) Descriptive epidemiology of Cornelia de Lange syndrome in Europe. Am J Med Genet A 146A:51–59
Beck B (1976) Epidemiology of Cornelia de Lange’s syndrome. Acta Paediatr Scand 65:631–638
Bhuiyan ZA, Klein M, Hammond P, van Haeringen A, Mannens MM, van Berckelaer-Onnes I, Hennekam RC (2006) Genotype–phenotype correlations of 39 patients with Cornelia De Lange syndrome: the Dutch experience. J Med Genet 43:568–575
Bhuiyan ZA, Stewart H, Redeker EJ, Mannens MM, Hennekam RC (2007) Large genomic rearrangements in NIPBL are infrequent in Cornelia de Lange syndrome. Eur J Hum Genet 15:505–508
Borck G, Redon R, Sanlaville D, Rio M, Prieur M, Lyonnet S, Vekemans M, Carter NP, Munnich A, Colleaux L, Cormier-Daire V (2004) NIPBL mutations and genetic heterogeneity in Cornelia de Lange syndrome. J Med Genet 41:e128
Bose T, Gerton JL (2010) Cohesinopathies, gene expression, and chromatin organization. J Cell Biol 189:201–210
Deardorff MA, Kaur M, Yaeger D, Rampuria A, Korolev S, Pie J, Gil-Rodríguez C, Arnedo M, Loeys B, Kline AD, Wilson M, Lillquist K, Siu V, Ramos FJ, Musio A, Jackson LS, Dorsett D, Krantz ID (2007) Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of Cornelia de Lange syndrome with predominant mental retardation. Am J Hum Genet 80:485–494
Deardorff MA, Wilde JJ, Albrecht M, Dickinson E, Tennstedt S, Braunholz D, Mönnich M, Yan Y, Xu W, Gil-Rodríguez MC, Clark D, Hakonarson H, Halbach S, Michelis LD, Rampuria A, Rossier E, Spranger S, Van Maldergem L, Lynch SA, Gillessen-Kaesbach G, Lüdecke HJ, Ramsay RG, McKay MJ, Krantz ID, Xu H, Horsfield JA, Kaiser FJ (2012a) RAD21 mutations cause a human cohesinopathy. Am J Hum Genet 90:1014–1027
Deardorff MA, Bando M, Nakato R, Watrin E, Itoh T, Minamino M, Saitoh K, Komata M, Katou Y, Clark D, Cole KE, De Baere E, Decroos C, Di Donato N, Ernst S, Francey LJ, Gyftodimou Y, Hirashima K, Hullings M, Ishikawa Y, Jaulin C, Kaur M, Kiyono T, Lombardi PM, Magnaghi-Jaulin L, Mortier GR, Nozaki N, Petersen MB, Seimiya H, Siu VM, Suzuki Y, Takagaki K, Wilde JJ, Willems PJ, Prigent C, Gillessen-Kaesbach G, Christianson DW, Kaiser FJ, Jackson LG, Hirota T, Krantz ID, Shirahige K (2012b) HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature 489:313–317
DeScipio C, Kaur M, Yaeger D, Innis JW, Spinner NB, Jackson LG, Krantz ID (2005) Chromosome rearrangements in Cornelia de Lange syndrome (CdLS): report of a der(3)t(3;12)(p25.3;p13.3) in two half sibs with features of CdLS and review of reported CdLS cases with chromosome rearrangements. Am J Med Genet A 137A:276–282
Dorsett D (2011) Cohesin: genomic insights into controlling gene transcription and development. Curr Opin Genet Dev 21:199–206
Dorsett D, Krantz ID (2009) On the molecular etiology of Cornelia de Lange syndrome. Ann N Y Acad Sci 1151:22–37
Feeney KM, Wasson CW, Parish JL (2010) Cohesin: a regulator of genome integrity and gene expression. Biochem J 428:147–161
Gillis LA, McCallum J, Kaur M, DeScipio C, Yaeger D, Mariani A, Kline AD, Li HH, Devoto M, Jackson LG, Krantz ID (2004) NIPBL mutational analysis in 120 individuals with Cornelia de Lange syndrome and evaluation of genotype–phenotype correlations. Am J Hum Genet 75:610–623
Kline AD, Krantz ID, Sommer A, Kliewer M, Jackson LG, FitzPatrick DR, Levin AV, Selicorni A (2007) Cornelia de Lange syndrome: clinical review, diagnostic and scoring systems, and anticipatory guidance. Am J Med Genet A 143A:1287–1296
Krantz ID, McCallum J, DeScipio C, Kaur M, Gillis LA, Yaeger D, Jukofsky L, Wasserman N, Bottani A, Morris CA, Nowaczyk MJ, Toriello H, Bamshad MJ, Carey JC, Rappaport E, Kawauchi S, Lander AD, Calof AL, Li HH, Devoto M, Jackson LG (2004) Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat Genet 36:631–635
Liu J, Zhang Z, Bando M, Itoh T, Deardorff MA, Clark D, Kaur M, Tandy S, Kondoh T, Rappaport E, Spinner NB, Vega H, Jackson LG, Shirahige K, Krantz ID (2009) Transcriptional dysregulation in NIPBL and cohesin mutant human cells. PLoS Biol 7:e1000119
Miyake N, Visser R, Kinoshita A, Yoshiura K, Niikawa N, Kondoh T, Matsumoto N, Harada N, Okamoto N, Sonoda T, Naritomi K, Kaname T, Chinen Y, Tonoki H, Kurosawa K (2005) Four novel NIPBL mutations in Japanese patients with Cornelia de Lange syndrome. Am J Med Genet A 135:103–105
Murray JE, Walayat M, Gillett P, Sharkey FH, Rajan D, Carter NP, Fitzpatrick DR (2012) An atypical facial appearance and growth pattern in a child with Cornelia de lange Syndrome: an intragenic deletion predicting loss of the N-terminal region of NIPBL. Clin Dysmorphol 21:22–23
Musio A, Selicorni A, Focarelli ML, Gervasini C, Milani D, Russo S, Vezzoni P, Larizza L (2006) X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nat Genet 38:528–530
Oliveira J, Dias C, Redeker E, Costa E, Silva J, Reis Lima M, den Dunnen JT, Santos R (2010) Development of NIPBL locus-specific database using LOVD: from novel mutations to further genotype–phenotype correlations in Cornelia de Lange Syndrome. Hum Mutat 31:1216–1222
Oliver C, Bedeschi MF, Blagowidow N, Carrico CS, Cereda A, Fitzpatrick DR, Gervasini C, Griffith GM, Kline AD, Marchisio P, Moss J, Ramos FJ, Selicorni A, Tunnicliffe P, Wierzba J, Hennekam RC (2010) Cornelia de Lange syndrome: extending the physical and psychological phenotype. Am J Med Genet A 152A:1127–1135
Pehlivan D, Hullings M, Carvalho CM, Gonzaga-Jauregui CG, Loy E, Jackson LG, Krantz ID, Deardorff MA, Lupski JR (2012) NIPBL rearrangements in Cornelia de Lange syndrome: evidence for replicative mechanism and genotype–phenotype correlation. Genet Med 14:313–322
Pié J, Gil-Rodríguez MC, Ciero M, López-Viñas E, Ribate MP, Arnedo M, Deardorff MA, Puisac B, Legarreta J, de Karam JC, Rubio E, Bueno I, Baldellou A, Calvo MT, Casals N, Olivares JL, Losada A, Hegardt FG, Krantz ID, Gómez-Puertas P, Ramos FJ (2010) Mutations and variants in the cohesion factor genes NIPBL, SMC1A, and SMC3 in a cohort of 30 unrelated patients with Cornelia de Lange syndrome. Am J Med Genet A 152A:924–929
Ratajska M, Wierzba J, Pehlivan D, Xia Z, Brundage EK, Cheung SW, Stankiewicz P, Lupski JR, Limon J (2010) Cornelia de Lange syndrome case due to genomic rearrangements including NIPBL. Eur J Med Genet 53:378–382
Russell KL, Ming JE, Patel K, Jukofsky L, Magnusson M, Krantz ID (2001) Dominant paternal transmission of Cornelia de Lange syndrome: a new case and review of 25 previously reported familial recurrences. Am J Med Genet 104:267–276
Russo S, Masciadri M, Gervasini C, Azzollini J, Cereda A, Zampino G, Haas O, Scarano G, Di Rocco M, Finelli P, Tenconi R, Selicorni A, Larizza L (2012) Intragenic and large NIPBL rearrangements revealed by MLPA in Cornelia de Lange patients. Eur J Hum Genet 20:734–741
Schoumans J, Wincent J, Barbaro M, Djureinovic T, Maguire P, Forsberg L, Staaf J, Thuresson AC, Borg A, Nordgren A, Malm G, Anderlid BM (2007) Comprehensive mutational analysis of a cohort of Swedish Cornelia de Lange syndrome patients. Eur J Hum Genet 15:143–149
Selicorni A, Russo S, Gervasini C, Castronovo P, Milani D, Cavalleri F, Bentivegna A, Masciadri M, Domi A, Divizia MT, Sforzini C, Tarantino E, Memo L, Scarano G, Larizza L (2007) Clinical score of 62 Italian patients with Cornelia de Lange syndrome and correlations with the presence and type of NIPBL mutation. Clin Genet 72:98–108
Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T (2004) NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet 36:636–641
Wierzba J, Kuzniacka A, Ratajska M, Lipska BS, Kardas I, Iliszko M, Limon J (2011) Cornelia de Lange syndrome associated with a de-novo novel NIPBL splice-site mutation and a coincidental inherited translocation t(3;5)(p13;q11). Clin Dysmorphol 20:222–224
Yan J, Saifi GM, Wierzba TH, Withers M, Bien-Willner GA, Limon J, Stankiewicz P, Lupski JR, Wierzba J (2006) Mutational and genotype–phenotype correlation analyses in 28 Polish patients with Cornelia de Lange syndrome. Am J Med Genet A 140:1531–1541
Zhang B, Chang J, Fu M, Huang J, Kashyap R, Salavaggione E, Jain S, Kulkarni S, Deardorff MA, Uzielli ML, Dorsett D, Beebe DC, Jay PY, Heuckeroth RO, Krantz I, Milbrandt J (2009) Dosage effects of cohesin regulatory factor PDS5 on mammalian development: implications for cohesinopathies. PLoS One 4:e5232
Acknowledgments
We thank the families and the collaborating physicians for their cooperation in this study.
This work has been financed by the Polish Ministry of Science and Higher Education grant N407 020 32/0530.
Author information
Authors and Affiliations
Corresponding author
Additional information
Alina Kuzniacka and Jolanta Wierzba contributed equally to this work.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Kuzniacka, A., Wierzba, J., Ratajska, M. et al. Spectrum of NIPBL gene mutations in Polish patients with Cornelia de Lange syndrome. J Appl Genetics 54, 27–33 (2013). https://doi.org/10.1007/s13353-012-0126-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-012-0126-9