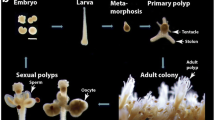
Abstract
The need for nonmammalian and nonvertebrate experimental systems for toxicology is increasing because of growing concern about animal welfare. Hydra, a genus of freshwater cnidarian, has been used in classical toxicology for more than 40 years and has been recommended as a standard model for ecotoxicology. Functional assays using Hydra homologues of the signal molecules and transcription factors of other metazoans have demonstrated that Hydra shares the common molecular mechanisms for pattern formation with bilaterian animals. The availability of the Hydra genome and mRNA information has meant that it can be used as a model organism in biological sciences. The purpose of this review is to explore recent developments in our understanding of the molecular aspects of axis formation, bud formation, regeneration, and the nervous system in Hydra and to introduce Hydra as an alternative animal model in environmental toxicogenomics for predicting the toxicity of environmental pollutants at the molecular level.
Similar content being viewed by others
References
Cowan, T. The Animal Welfare Act: Background and Selected Animal Welfare Legislation. Congressional Research Service 7-5700 (2013).
Galliot, B. Hydra, a fruitful model system for 270 years. Int J Dev Biol 56:411–423 (2012).
Pollino, C. A. & Holdway, D. A. Potential of two Hydra species as standard toxicity test animals. Ecotoxicol Environ Saf 43:309–316 (1999).
Snape, J. R., Maund, S. J., Pickford, D. B. & Hutchinson, T. H. Ecotoxicogenomics: the challenge of integrating genomics into aquatic and terrestrial ecotoxicology. Aquat Toxicol 67:143–154 (2004).
Bode, H. R. The head organizer in Hydra. Int J Dev Biol 56:473–478 (2012).
Hobmayer, B. et al. WNT signalling molecules act in axis formation in the diploblastic metazoan Hydra. Nature 407:186–189 (2000).
Chapman, J. A. et al. The dynamic genome of Hydra. Nature 464:592–596 (2010).
Medina, M., Collins, A. G., Silberman, J. D. & Sogin, M. L. Evaluating hypotheses of basal animal phylogeny using complete sequences of large and small subunit rRNA. Proc Natl Acad Sci USA 98:9707–9712 (2001).
Kim, J., Kim, W. & Cunningham, C. W. A new perspective on lower metazoan relationships from 18S rDNA sequences. Mol Biol Evol 16:423–427 (1999).
Wainright, P. O., Hinkle, G., Sogin, M. L. & Stickel, S. K. Monophyletic origins of the metazoa: an evolutionary link with fungi. Science 260:340–342 (1993).
Quinn, B., Gagne, F. & Blaise, C. Hydra, a model system for environmental studies. Int J Dev Biol 56:613–625 (2012).
Lengfeld, T. et al. Multiple Wnts are involved in Hydra organizer formation and regeneration. Dev Biol 330:186–199 (2009).
Guder, C. et al. An ancient Wnt-Dickkopf antagonism in Hydra. Development 133:901–911 (2006).
De Robertis, E. M., Larrain, J., Oelgeschlager, M. and Wessely, O. The establishment of Spemann’s organizer and patterning of the vertebrate embryo. Nat Rev Genet 1:171–181 (2000).
Broun, M., Gee, L., Reinhardt, B. & Bode, H. R. Formation of the head organizer in Hydra involves the canonical Wnt pathway. Development 132:2907–2916 (2005).
Nakamura, Y., Tsiairis, C. D., Ozbek, S. & Holstein, T. W. Autoregulatory and repressive inputs localize Hydra Wnt3 to the head organizer. Proc Natl Acad Sci USA 108:9137–9142 (2011).
Johnson, E. M. & Gabel, B. E. An artificial ‘embryo’ for detection of abnormal developmental biology. Fundam Appl Toxicol 3:243–249 (1983).
Quinn, B., Gagne, F. & Blaise, C. Evaluation of the acute, chronic and teratogenic effects of a mixture of eleven pharmaceuticals on the cnidarian, Hydra attenuata. Sci Total Environ 407:1072–1079 (2009).
Johnson, E. M., Gorman, R. M., Gabel, B. E. & George, M. E. The Hydra attenuata system for detection of teratogenic hazards. Teratog Carcinog Mutagen 2:263–276 (1982).
Wilby, O. K. & Tesh, J. M. The Hydra assay as an early screen for teratogenic potential. Toxicol In Vitro 4:582–583 (1990).
Brooun, M. et al. Organizer formation in Hydra is disrupted by thalidomide treatment. Dev Biol 378:51–63 (2013).
Otto, J. J. & Campbell, R. D. Budding in Hydra attenuata: bud stages and fate map. J Exp Zool 200:417–428 (1977).
Mitchell, F. M. & Holdway, D. A. The acute and chromic toxicity of the dispersants Corexit 9527 and 9500, water accommodated fraction (WAF) of crude oil, and dispersant enhanced WAF (DEWAF) to Hydra viridissima (green hydra). Wat Res 34:343–348 (2000).
Bielen, H. et al. Divergent functions of two ancient Hydra Brachyury paralogues suggest specific roles for their C-terminal domains in tissue fate induction. Development 134:4187–4197 (2007).
Technau, U. & Bode, H. R. HyBra1, a Brachyury homologue, acts during head formation in Hydra. Development 126:999–1010 (1999).
Fujisawa, T. Hydra regeneration and epitheliopeptides. Dev Dyn 226:182–189 (2003).
Lohmann, J. U. & Bosch, T. C. The novel peptide HEADY specifies apical fate in a simple radially symmetric metazoan. Genes Dev 14:2771–2777 (2000).
Takahashi, T. et al. Hym-301, a novel peptide, regulates the number of tentacles formed in Hydra. Development 132:2225–2234 (2005).
Hoffmeister, S. A. Isolation and characterization of two new morphogenetically active peptides from Hydra vulgaris. Development 122:1941–1948 (1996).
Grens, A. et al. The novel signal peptides, pedibin and Hym-346, lower positional value thereby enhancing foot formation in Hydra. Development 126:517–524 (1999).
Hoffmeister-Ullerich, S. A. The foot formation stimulating peptide pedibin is also involved in patterning of the head in Hydra. Mech Dev 106:37–45 (2001).
Harafuji, N. et al. Enhancement of foot formation in Hydra by a novel epitheliopeptide, Hym-323. Development 128:437–446 (2001).
Amimoto, Y., Kodama, R. & Kobayakawa, Y. Foot formation in Hydra: a novel gene, anklet, is involved in basal disk formation. Mech Dev 123:352–361 (2006).
Cardenas, M. et al. Selective protein kinase inhibitors block head-specific differentiation in Hydra. Cell Signal 12:649–658 (2000).
Takahashi, T. et al. Systematic isolation of peptide signal molecules regulating development in Hydra: LWa-mide and PW families. Proc Natl Acad Sci USA 94:1241–1246 (1997).
Fujisawa, T. Hydra peptide project 1993–2007. Dev Growth Differ 50 Suppl 1:S257–268 (2008).
Yum, S. et al. A novel neuropeptide, Hym-176, induces contraction of the ectodermal muscle in Hydra. Biochem Biophys Res Commun 248:584–590 (1998).
Yum, S., Takahashi, T., Hatta, M. & Fujisawa, T. The structure and expression of a preprohormone of a neuropeptide, Hym-176 in Hydra magnipapillata. FEBS Lett 439:31–34 (1998).
Takahashi, T. et al. A novel neuropeptide, Hym-355, positively regulates neuron differentiation in Hydra. Development 127:997–1005 (2000).
Woo, S. et al. Toxaphene affects the levels of mRNA transcripts that encode antioxidant enzymes in Hydra. Comp Biochem Physiol C Toxicol Pharmacol 156:37–41 (2012).
Takahashi, T. et al. Identification of a new member of the GLWamide peptide family: physiological activity and cellular localization in cnidarian polyps. Comp Biochem Physiol B Biochem Mol Biol 135:309–324 (2003).
Hayakawa, E., Takahashi, T., Nishimiya-Fujisawa, C. & Fujisawa, T. A novel neuropeptide (FRamide) family identified by a peptidomic approach in Hydra magnipapillata. FEBS J 274:5438–5448 (2007).
Takahashi, T. & Hamaue, N. Molecular characterization of Hydra acetylcholinesterase and its catalytic activity. FEBS Lett 584:511–516 (2010).
Dash, B. et al. Molecular characterization of two superoxide dismutases from Hydra vulgaris. Gene 387:93–108 (2007).
Dash, B. et al. Molecular characterization of phospholipid hydroperoxide glutathione peroxidases from Hydra vulgaris. Gene 381:1–12 (2006).
Dash, B. & Phillips, T. D. Molecular characterization of a catalase from Hydra vulgaris. Gene 501:144–152 (2012).
Ankley, G. T. et al. Toxicogenomics in regulatory ecotoxicology. Environ Sci Technol 40:4055–4065 (2006).
Author information
Authors and Affiliations
Corresponding author
Additional information
These authors contributed equally to this work.
Rights and permissions
About this article
Cite this article
Yum, S., Woo, S., Lee, A. et al. Hydra, a candidate for an alternative model in environmental genomics. Mol. Cell. Toxicol. 10, 339–346 (2014). https://doi.org/10.1007/s13273-014-0038-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13273-014-0038-3