Abstract
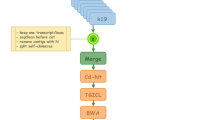
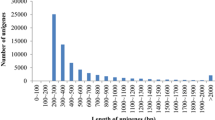
In this study, the whole transcriptome of the Pacific oyster Crassostrea gigas was sequenced using Illumina RNA-seq. De novo assembly was performed with 33,750,764 raw reads using Trinity, which assembled 87,887 contigs. Transdecoder found 41,542 candidate coding contigs which showed homology to other species by BLAST analysis. Functional gene annotation was performed by Gene Ontology and KEGG pathway analyses. Finally, we identified a number of expressed gene pathways for C. gigas representing a useful model animal for gene information-based study such as environmental monitoring, immune-relevant aquaculture, and ecotoxicogenomics studies to uncover molecular mechanisms of stress-triggered sensitivity and physiological response to C. gigas.



Similar content being viewed by others
References
Akira S, Takeda K (2004) Toll-like receptor signalling. Nat Rev Immunol 4:499–511
Alzieu C, Heral M (1984) Ecotoxicological effects of organotin compounds on oyster culture. In: Persoone G, Jaspers E, Claus C (eds) Ecotoxicological testing for the marine environment, vol 2. State University, Belgium, pp 187–195
Bourlat SJ, Borja A, Gilbert J, Taylor MI, Davies N, Weisberg SB, Griffith JF, Lettieri T, Field D, Benzie J et al (2013) Genomics in marine monitoring: new opportunities for assessing marine health status. Mar Pollut Bull 74:19–31
Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Curole JP, Hedgecock D (2007) Bivalve genomics: complications, challenges, and future perspectives. In: Liu ZJ (ed) Aquaculture genome technologies. Blackwell Publishing, Ames, pp 525–543
de Lorgeril J, Zenagui R, Rosa RD, Piquemal D, Bachère E (2011) Whole transcriptome profiling of successful immune response to Vibrio infections in the oyster Crassostrea gigas by digital gene expression analysis. PLoS One 6:e23142
Du Y, Zhang L, Huang B, Guan X, Li L, Zhang G (2013) Molecular cloning, characterization, and expression of two myeloid differentiation factor 88 (Myd88) in Pacific oyster, Crassostrea gigas. J World Aquac Soc 44:759–774
Feldmeyer B, Wheat CW, Krezdorn N, Rotter B, Pfenninger M (2011) Short read Illumina data for the de novo assembly of a non-model snail species transcriptome (Radix balthica, Basommatophora, Pulmonata), and a comparison of assembler performance. BMC Genomics 12:317
Fent K, Sumpter JP (2011) Progress and promises in toxicogenomics in aquatic toxicology: is technical innovation driving scientific innovation? Aquat Toxicol 105:25–39
Food and Agriculture Organization (FAO) (2015) World aquaculture production of fish, crustaceans, molluscs, etc., by principal species in 2013
Forrest BM, Keeley NB, Hopkins GA, Webb SC, Clement DM (2009) Bivalve aquaculture inestuaries: review and synthesis of oyster cultivation effects. Aquaculture 298:1–15
Gees M, Colsoul B, Nilius B (2010) The role of transient receptor potential cation channels in Ca2+ signaling. Cold Spring Harb Perspect Biol 2:a003962
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q et al (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652
Hedgecock D, Gaffney PM, Goulletquer P, Guo X, Reece K, Warr GW (2005) The case for sequencing the Pacific oyster genome. J Shellfish Res 24:429–441
Ho KK, Leung PT, Ip JC, Qiu JW, Leung KM (2014) De novo transcriptomic profile in the gonadal tissues of the intertidal whelk Reishia clavigera. Mar Pollut Bull 85:499–504
Hubert S, Hedgecock D (2004) Linkage maps of microsatellite DNA markers for the Pacific oyster Crassostrea gigas. Genetics 168:351–362
Malham SK, Cotter E, O’Keeffe S, Lynch S, Culloty SC, King JW, Latchford JW, Beaumont AR (2009) Summer mortality of the Pacific oyster, Crassostrea gigas, in the Irish Sea: the influence of temperature and nutrients on health and survival. Aquaculture 287:128–138
Montagnani C, Kappler C, Reichhart JM, Escoubas JM (2004) Cg-Rel, the first Rel/NF-κB homolog characterized in a mollusk, the Pacific oyster Crassostrea gigas. FEBS Lett 561:75–82
Newell RIE (2004) Ecosystem influences of natural and cultivated populations of suspension-feeding bivalve molluscs: a review. J Shellfish Res 23:51–61
Nilius B, Owsianik G (2011) The transient receptor potential family of ion channels. Genome Biol 12:218
Ponder WF, Colgan DJ, Healy JM, Nützel A, Simone LRL, Strong EE (2008) Caenogastropoda. In: Ponder WF, Lindberg DL (eds) Molluscan phylogeny and evolution. University of California Press, Berkeley, pp 331–383
Quayle DB, Newkirk GF (1989) Farming bivalve molluscs: methods for study and development. World Aquaculture Society, Baton Rouge
Riesgo A, Andrade SC, Sharma PP, Novo M, Pérez-Porro AR, Vahtera V, González VL, Kawauchi GY, Giribet G (2012) Comparative description of ten transcriptomes of newly sequenced invertebrates and efficiency estimation of genomic sampling in non-model taxa. Front Zool 9:33
Sadamoto H, Takahashi H, Okada T, Kenmoku H, Toyota M, Asakawa Y (2012) De novo sequencing and transcriptome analysis of the central nervous system of mollusc Lymnaea stagnalis by deep RNA sequencing. PLoS One 7:e42546
Soletchnik P, Ropert M, Mazurié J, Fleury PG, Le Coz F (2007) Relationships between oyster mortality patterns and environmental data from monitoring databases along the coasts of France. Aquaculture 271:384–400
Tirapé A, Bacque C, Brizard R, Vandenbulcke F, Boulo V (2007) Expression of immune-related genes in the oyster Crassostrea gigas during ontogenesis. Dev Comp Immunol 31:859–873
Tong Y, Zhang Y, Huang J, Xiao S, Zhang Y, Li J, Chen J, Yu Z (2015) Transcriptomics analysis of Crassostrea hongkongensis for the discovery of reproduction-related genes. PLoS One 10:e0134280
Yum S, Woo S, Lee A, Won H, Kim J (2014) Hydra, a candidate for an alternative model in environmental genomics. Mol Cell Toxicol 10:339–346
Zhang G, Fang X, Guo X, Li L, Luo R, Xu F, Yang P, Zhang L, Wang X, Qi H et al (2012) The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490:49–54
Zhang Y, He X, Yu F, Xiang Z, Li J, Thorpe KL, Yu Z (2013) Characteristic and functional analysis of toll-like receptors (TLRs) in the lophotrocozoan, Crassostrea gigas, reveals ancient origin of TLR-mediated innate immunity. PLoS One 8:e76464
Zhang L, Li L, Zhu Y, Zhang G, Guo X (2014) Transcriptome analysis reveals a rich gene set related to innate immunity in the Eastern oyster (Crassostrea virginica). Mar Biotechnol 16:17–33
Zhang L, Li L, Guo X, Litman GW, Dishaw LJ, Zhang G (2015) Massive expansion and functional divergence of innate immune genes in a protostome. Sci Rep 5:8693
Zhao X, Yu H, Kong L, Liu S, Li Q (2014) Comparative transcriptome analysis of two oysters, Crassostrea gigas and Crassostrea hongkongensis provides insights into adaptation to hypo-osmotic conditions. PLoS One 9:e111915
Acknowledgments
This work was supported by a Grant (RP-2015-AG-089) funded to Hyun-Jeong Lim, National Institute of Fisheries Science of the Korean government.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Compliance of bioethical standard
All the experiments were approved by the animal care and use committee of National Fisheries Research and Development Institute (NFRDI, Pusan, South Korea).
Conflict of Interest
All authors claim no conflicts of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lim, HJ., Lim, JS., Lee, JS. et al. Transcriptome profiling of the Pacific oyster Crassostrea gigas by Illumina RNA-seq. Genes Genom 38, 359–365 (2016). https://doi.org/10.1007/s13258-015-0376-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-015-0376-0