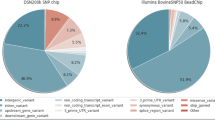
Abstract
Korean native cattle, known as Hanwoo, have been raised in the Korean Peninsula since 2000 B.C. for use as a draft animal. However, Hanwoo now have an important position in the Korean livestock industry as a meat source. Therefore, the breeding and selection of Hanwoo are crucial for the industry. Although many researchers have studied the genetic architecture of Hanwoo, no well-established Hanwoo-related databases exist. In order to better understand the genetic contents of Hanwoo, an integrated database is necessary. We constructed a genetic variants database including annotation information. HanwooGDB (http://hanwoogdb.snu.ac.kr) provides genetic variants (SNPs, INDELs, CNVs) in the Hanwoo genome produced by Next Generation Sequencing data collected from 23 cattle. The identified SNPs were integrated with SNP chip data and annotation information for checking the concordance of position of each SNP and inferring functional aspects. This database provides browsers to understand and visualize the comprehensive information of these variants and allows users to download data according to their preference from this database without limitation. This database will contribute to genetic research and development of Hanwoo breeding strategies.



Similar content being viewed by others
References
Browning SR, Browning BL (2007) Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet 81:1084–1097
Cheong HS, Kim LH, Namgoong S, Shin HD (2013) Development of discrimination SNP markers for Hanwoo (Korean native cattle). Meat Sci 94:355–359
Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6:80–92
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST (2011) The variant call format and VCFtools. Bioinformatics 27:2156–2158
Edea Z, Dadi H, Kim SW, Dessie T, Kim KS (2012) Comparison of SNP variation and distribution in indigenous Ethiopian and Korean Cattle (Hanwoo) populations. Genomics Inform 10:200–205
Excoffier L, Lischer HE (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S (2012) Ensembl 2012. Nucleic Acids Res 40:D84–D90
Handsaker RE, Korn JM, Nemesh J, McCarroll SA (2011) Discovery and genotyping of genome structural polymorphism by sequencing on a population scale. Nat Genet 43:269–276
Hwang YH, Kim GD, Jeong JY, Hur SJ, Joo ST (2010) The relationship between muscle fiber characteristics and meat quality traits of highly marbled Hanwoo (Korean native cattle) steers. Meat Sci 86:456–461
Jiang J, Jiang L, Zhou B, Fu W, Liu JF, Zhang Q (2011) Snat: a SNP annotation tool for bovine by integrating various sources of genomic information. BMC Genet 12:85
Jo C, Cho SH, Chang J, Nam KC (2012) Keys to production and processing of Hanwoo beef: a perspective of tradition and science. Anim Front 2:32–38
Kim JB, Lee C (2000) Historical look at the genetic improvement in Korean cattle. Rev Asian Australas J Anim Sci 13:1467–1481
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359
Lee SH, Park EW, Cho YM, Kim SK, Lee JH, Jeon JT, Lee CS, Im SK, Oh SJ, Thompson JM (2007) Identification of differentially expressed genes related to intramuscular fat development in the early and late fattening stages of hanwoo steers. J Biochem Mol Biol 40:757–764
Lee SH, Cho YM, Lee SH, Kim BS, Kim NK, Choy YH, Kim KH, Yoon DH, Im SK, Oh SJ et al (2008a) Identification of marbling-related candidate genes in M. longissimus dorsi of high- and low marbled Hanwoo (Korean Native Cattle) steers. BMB Rep 41:846–851
Lee YS, Lee JH, Lee JY, Kim JJ, Park HS, Yeo JS (2008b) Identification of candidate SNP (single nucleotide polymorphism) for growth and carcass traits related to QTL on chromosome 6 in Hanwoo (Korean cattle). Asian Aust J Anim Sci 21:1703–1709
Lee YM, Han CM, Li Yi, Lee JJ, Kim LH, Kim JH, Kim DI, Lee SS, Park BL, Shin HD (2010) A whole genome association study to detect single nucleotide polymorphism for carcass traits in Hanwoo populations. Asian Australas J Anim Sci 23:417–424
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303
Reese JT, Childers CP, Sundaram JP, Dickens CM, Childs KL, Vile DC, Elsik CG (2010) Bovine Genome Database: supporting community annotation and analysis of the Bos taurus genome. BMC Genomics 11:645
Shin SC, Heo JP, Chung ER (2012) Genetic variants of the FABP4 gene are associated with marbling scores and meat quality grades in Hanwoo (Korean cattle). Mol Biol Rep 39:5323–5330
Acknowledgments
This study was supported by grants from Research Program (Project No: PJ008487) of the National Livestock Research Institute and Next Generation BioGreen 21 Program (Project No: PJ008191 and PJ00995401) of Rural Development Administration, Republic of Korea.
Conflict of Interest
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Kwondo Kim and Woori Kwak have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Kim, K., Kwak, W., Sung, SS. et al. A novel genetic variant database for Korean native cattle (Hanwoo): HanwooGDB. Genes Genom 37, 15–22 (2015). https://doi.org/10.1007/s13258-014-0224-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-014-0224-7