Abstract
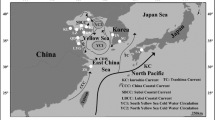
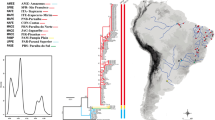
The native Asian oyster, Crassostrea ariakensis is one of the most common and important Crassostrea species that occur naturally along the coast of East Asia. Molecular species diagnosis is a prerequisite for population genetic analysis of wild oyster populations because oyster species cannot be discriminated reliably using external morphological characters alone due to character ambiguity. To date there have been few phylogeographic studies of natural edible oyster populations in East Asia, in particular this is true of the common species in Korea C. ariakensis. We therefore assessed the levels and patterns of molecular genetic variation in East Asian wild populations of C. ariakensis from Korea, Japan, and China using DNA sequence analysis of five concatenated mtDNA regions namely; 16S rRNA, cytochrome oxidase I, cytochrome oxidase II, cytochrome oxidase III, and cytochrome b. Two divergent C. ariakensis clades were identified between southern China and remaining sites from the northern region. In addition, hierarchical AMOVA and pairwise Φ ST analyses showed that genetic diversity was discontinuous among wild populations of C. ariakensis in East Asia. Biogeographical and historical sea level changes are discussed as potential factors that may have influenced the genetic heterogeneity of wild C. ariakensis stocks across this region.




Similar content being viewed by others
References
An YK, Yoon HS, Choi SD (2006) Effects of temperature, salinity on the growth of Crassostrea ariakensis in Seomjin River. Korean J Environ Biol 24:60–66
An HS, Lee JW, Park JY, Jung HT (2013) Genetic structure of the Korean black scraper Thamnaconus modestus inferred from microsatellite marker analysis. Mar Biol Rep 40:3445–3456
Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48
Bushek D, Kornbluh A, Debrosse G, Wang H, Guo X (2006) Oyster sex wars: evidence for a gamete sink if Crassostrea virginica and Crassostrea ariakensis spawn synchronously. J Shellfish Res 25:716
Cahn AR (1950) Oyster culture in Japan (Fishery leaflet). Fish and Wildlife Service, US
Cho YK, Seo GH, Kim CS, Choi BK, Shaha DC (2013) Role of wind stress in causing maximum transport through the Korea Strait in autumn. J Mar Syst 115–116:33–39
Coan EV, Scott PV, Bernard FR (2000) Bivalve seashells of western North America: marine bivalve mollusks from Arctic Alaska to Baja California. Santa Barbara Museum of Natural History, Santa Barbara
Cordes JF, Xiao J, Reece KS (2008) Discrimination of nine Crassostrea oyster species based upon restriction fragment-length polymorphism analysis of nuclear and mitochondrial DNA markers. J Shellfish Res 27:1155–1161
DeWoody JA, Avise JC (2000) Microsatellite variation in marine, freshwater and an adromous fishes compared with other animals. J Fish Biol 56:461–473
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Excoffier L, Larval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Fleury E, Huvet A, Lelong C, de Lorgeri J, Boulo V, Gueguen Y et al (2009) Generation and analysis of a 29,745 unique expressed sequence tags from the Pacific oyster (Crassostrea gigas) assembled into a publicly accessible database: the Gigas Database. BMC Genom 10:341
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3:294–299
Fu YX (1997) Statistical tests for neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925
Guo X, Zhang G, Qian L, Wang H, Liu X, Wang A (2006) Oysters and oyster farming in China: a review. J Shellfish Res 25:734
Heads M (2005) Towards a panbiogeography of the seas. Biol J Linn Socy 84:675–723
Hubert S, Hedgecock D (2004) Linkage maps of microsatellite DNA markers for the Pacific oyster Crassostrea gigas. Genetics 168:351–362
Jung H, Kim WJ, Gaffney PM (2006) Development of single nucleotide polymorphisms (SNPs) in Crassostrea ariakensis and related Crassostrea species. J Shellfish Res 25:742
Kang JH, Park JY, Choi TJ (2012) Genetic differentiation of octopuses from different habitats near the Korean Peninsula and eastern China based on analysis of the mDNA cytochrome C oxidase 1 gene. Genet Mol Res 11:3988–3997
Kim WJ, Lee JH, Kim KK, Kim YO, Nam BH, Kong HJ, Jung HT (2009) Genetic relationships of four Korean oysters based on RAPD and nuclear rDNA ITS sequence analyses. Korean J Malacol 25:41–49
Kim WJ, Jung H, Gaffney PM (2011) Development of type I genetic markers from expressed sequence tags in highly polymorphic species. Mar Biotechnol 13:127–132
Kwan YS, Song HK, Lee HJ, Lee WO, Won YJ (2012) Population genetic structure and evidence of demographic expansion of the Ayu (Plecoglossus altivelis) in East Asia. Anim Syst Evol Div 28:279–290
Kwun HJ, Song YS, Myoung SH, Kim JK (2013) Two new records of juvenile Oedalechilus labiosus and Ellochelon vaigiensis (Mugiliformes: Mugilidae) from Jeju island, Korea, as revealed by molecular analysis. Fish Aquat Sci 16:109–116
Lambeck K, Esat TM, Potter EK (2002) Links between climate and sea levels for the past three million years. Nature 419:199–206
Lee SY, Park DW, An HS, Kim SH (2000) Phylogenetic relationship among four species of Korean oysters based on mitochondrial 16S rDNA and COI gene. Korean J Syst Zool 162:203–211
Lie HJ, Cho CH, Lee JH, Niller P, Hu JH (1998) Separation of the Kuroshio water and its penetration onto the continental shelf west of Kyushu. J Geophys Res 103:2963–2976
Liu JX, Gao TX, Yokogawa K, Zhang YP (2006) Differential population structuring and demographic history of two closely related fish species, Japanese Sea bass (Lateolabrax japonicus) and spotted sea bass (Lateolabrax maculatus) in Northwestern Pacific. Mol Phylogenet Evo 39:799–811
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Can Res 27:209–220
Milbury CA, Gaffney PM (2005) Complete mitochondrial DNA sequence of the eastern oyster Crassostrea virginica. Mar Biotech 7:697–712
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Obata M, Shimizu M, Sano N, Komaru A (2008) Maternal inheritance of mitochondrial DNA (mtDNA) in the Pacific oyster (Crassostrea gigas): a preliminary study using mtDNA sequence analysis with evidence of random distribution of MitoTracker-stained sperm mitochondira in fertilized eggs. Zool Sci 25:248–254
Pagel M, Meade A (2004) A phylogenetic mixture model for detecting pattern heterogenetity in gene sequence or character-state data. Syst Biol 53:571–581
Palumbi SR (1994) Genetic divergence, reproductive isolation, and marine speciation. Annu Rev Ecol Syst 25:547–572
Pang IC, Rho HK, Kim TH (1992) Seasonal variation of water mass distributions and their causes in the Yellow Sea, the East China Sea and the adjacent seas of Cheju Island. Bull Korean Fish Soc 25:151–163
Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818
Quilang J, Wang S, Li P, Abernathy J, Peatman E, Wang Y, Wang L, Shi Y, Wallace R, Guo X, Liu Z (2007) Generation and analysis of ESTs from the eastern oyster, Crassostrea virginica Gmelin and identification of microsatellite and SNP markers. BMC Genom 8:157
Reeb CA, Avise JC (1990) A genetic discontinuity in a continuously distributed species: mitochondrial DNA in the American oyster, Crassostrea virginica. Genetics 124:397–406
Reece KS, Cordes JF, Stubbs JB, Hudson KL, Francis EA (2008) Molecular phylogenies help resolve taxonomic confusion with Asian Crassostrea oyster species. Mar Biol 153:709–721
Ren J, Liu X, Jiang F, Guo X, Liu B (2010) Unusual conservation of mitochondrial gene order in Crassostrea oysters: evidence for recent speciation in Asia. BMC Evol Biol 10:394
Rice WJ (1989) Analyzing tables of statistical tests. Evolution 43:223–225
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Rozas J, Sanchez-Delbarro JC, Messegue X, Rozas R (2003) DnaSp, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497
Sekino M, Sato S, Hong JS, Li Q (2012) Contrasting pattern of mitochondrial population diversity between an estuarine bivalve, the Kumamoto oyster Crassostrea sikamea, and the closely related Pacific oyster C. gigas. Mar Biol 159:2757–2776
Smouse PE, Long JC, Sokal RR (1986) Multiple regression and correlation extensions of the Mantel test of matrix correspondence. System Zoo 35:627–632
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690
Tajima F (1983) Evolutionary relationship of DNA sequences in finite populations. Genetics 105:437–460
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Tamura K, Peterson D, Peterson N, Nei G, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thorson G (1950) Reproductive and larval ecology of marine bottom invertebrates. Biol Rev 25:1–45
Varney RL, Galindo-Sánchez CE, Cruz P, Gaffney PM (2009) Population genetics of the eastern oyster Crassostrea virginica (Gmelin, 1791) in the Gulf of Mexico. J Shellfish Res 28:855–864
Wang H, Guo X (2008a) Identification of Crassostrea ariakensis and related oysters by multiplex species-specific PCR. J Shellfish Res 27:481–488
Wang H, Guo X (2008b) ITS length polymorphism in oyster and its use in species identification. J Shellfish Res 27:489–494
Wang H, Guo X, Zhan G, Zhang F (2004) Classification of jinjiang oysters Crassostrea rivularis (Gould, 1861) from China, based on morphology and phylogenetic analysis. Aquaculture 242:137–155
Wang H, Zhang G, Liu X, Guo X (2008a) Classification of common oysters from North China. J Shellfish Res 27:495–504
Wang M, Zhang X, Yang T, Han Z, Yanagimoto T, Gao T (2008b) Genetic diversity in the mtDNA control region and population structure in the Sardinella zunasi Bleeker. African J Biotechnol 7:4384–4392
Watterson GA (1975) On the number of segregating sites in genetical models without recombination. Theor Pop Biol 7:256–276
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Wilberg MJ, Livings ME, Barkman JS, Morris BT, Robinson JM (2011) Overfishing, disease, habitat loss, and potential extirpation of oysters in upper Chesapeake Bay. Mar Ecol Prog Ser 436:131–144
Wu XY, Xu XD, Yu ZN, Wei ZP, Xia JJ (2010) Comparison of seven Crassostrea mitogenomes and phylogenetic analyses. Mol Phylogenet Evol 57:448–454
Xiao S, Li A, Jiang F, Li T, Wan S, Huang P (2004) The history of the Yangtze River entering sea since the Last Glacial Maxima: a review and look forward. J Coast Res 20:599–604
Xiao J, Cordes JF, Wang H, Guo X, Reece KS (2010) Population genetics of Crassostrea ariakensis in Asia inferred from microsatellite markers. Mar Biol 157:1767–1781
Xu F (1997) Bivalves from China Seas. Science Press, Beijing
Yang EC, Lee SY, Lee WJ, Boo SM (2009) Molecular evidence for recolonization of Ceramium japonicum (Ceramiaceae, Rhodophyta) on the west coast of Korea after the last glacial maximum. Bot Mar 52:307–315
Yoo SK, Lim HK, Jang YJ (2004) Growth and spawning of Crassostrea rivularis from the southern sea of Korea. Korean J Malacol 20:131–134
Yoon HS, Jung H, Choi SD (2008) Suminoe oyster (Crassostrea ariakensis) culture in Korea. J Shellfish Res 27:505–509
Yu Z, Guo X (2005) Genetic analysis of selected strains of eastern oyster (Crassostrea virginica Gmelin) using AFLP and microsatellite markers. Mar Biotechnol 6:575–586
Yu Z, Wei Z, Kong X, Shi W (2008) Complete mitochondrial DNA sequence of oyster Crassostrea hongkongensis: a case of “Tandem duplication random loss” for genome rearrangement in Crassostea? BMC Genom 9:477
Zhang Q, Allen SK, Reece KS (2005) Genetic variation in wild and hatchery stocks of Suminoe oyster (Crassostrea ariakensis) assessed by PCR-RFLP and microsatellite markers. Mar Biotech 7:1–13
Zhou M, Allen SK (2003) A review of published work on Crassostrea ariakensis. J Shellfish Res 22:1–20
Zhu J, Chen C, Ding P, Li C, Lin H (2004) Does the Taiwan warm current exist in winter? Geophys Res Lett 31:L12302
Acknowledgments
The authors would like to acknowledge the help provided by Dr. Peter Mather, Dr. David Hurwood and Dr. Patrick Gaffney in regard to constructive comments and valuable suggestions on the manuscript. The authors also thank Dr. Ximing Guo and Dr. Kimberly Reece for providing priceless samples from China and Japan, and Dr. Patrick Gaffney for sequencing of a few Chinese and Japanese samples. This study was supported by Korea Science and Engineering Foundation (Grant No. 2005-215-F00009), by a partial grant from the National Fisheries Research and Development Institute (RP-2014-BT-013), and by a partial grant from the Ministry of Maritime Affairs and Fisheries (Technical Development for Higher Productivity of Kang-gul), Korea.
Conflict of interest
The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Additional information
W.-J. Kim, S. T. Dammannagoda and H. Jung contributed equally to this work.
Rights and permissions
About this article
Cite this article
Kim, WJ., Dammannagoda, S.T., Jung, H. et al. Mitochondrial DNA sequence analysis from multiple gene fragments reveals genetic heterogeneity of Crassostrea ariakensis in East Asia. Genes Genom 36, 611–624 (2014). https://doi.org/10.1007/s13258-014-0198-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-014-0198-5