Abstract
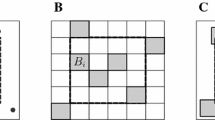
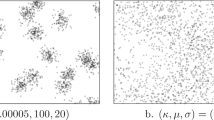
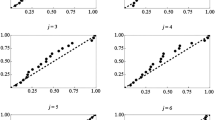
To estimate the spatial intensity (density) of plants and animals, ecologists often sample populations by prespecifing a spatial array of points, then measuring the distance from each point to the k nearest organisms, a so-called k-tree sampling method. A variety of ad hoc methods are available for estimating intensity from k-tree sampling data, but they assume that two distinct points of the array do not share nearest neighbors. However, nearest neighbors are likely to be shared when the population intensity is low, as it is in our application. The purpose of this paper is twofold: (a) to derive and use for estimation the likelihood function for a k-tree sample under an inhomogeneous Poisson point-process model and (b) to estimate spatial intensity when nearest neighbors are shared. We derive the likelihood function for an inhomogeneous Poisson point-process with intensity λ(x,y) and propose a likelihood-based, kernel-smoothed estimator \(\hat{\lambda}(x,y)\). Performance of the method for k=1 is tested on four types of simulated populations: two homogeneous populations with low and high intensity, a population simulated from a bivariate normal distribution of intensity, and a “cliff” population in which the region is divided into high- and low-intensity subregions. The method correctly detected spatial variation in intensity across different subregions of the simulated populations. Application to 1-tree samples of carnivorous pitcher plants populations in four New England peat bogs suggests that the method adequately captures empirical patterns of spatial intensity. However, our method suffers from two evident sources of bias. First, like other kernel smoothers, it underestimates peaks and overestimates valleys. Second, it has positive bias analogous to that of the MLE for the rate parameter of exponential random variables.
Similar content being viewed by others
References
Augustin, N. H., Musio, M., von Wilpert, K., Kublin, E., Wood, S. N., and Schumacher, M. (2009), “Modeling Spatiotemporal Forest Health Monitoring Data,” Journal of the American Statistical Association, 104, 899–911.
Barbour, M. C., Burk, J. H., Pitts, W. D., Gilliam, F. S., and Schwartz, M. W. (1999), Terrestrial Plant Ecology (3rd ed.), Menlo Park: Benjamin Cummings.
Byth, K., and Ripley, B. D. (1980), “On Sampling Spatial Patterns by Distance Methods,” Biometrics, 36, 279–284.
Damggard, C. (2009), “On the Distribution of Plant Abundance Data,” Ecological Informatics, 4, 76–82.
Diggle, P. J. (1975), “Robust Density Estimation Using Distance Methods,” Biometrika, 62, 39–48.
— (1977), “A Note on Robust Density Estimation for Spatial Point Patterns,” Biometrika, 64, 91–95.
Ellison, A. M., and Parker, J. N. (2002), “Seed Dispersal and Seedling Establishment of Sarracenia Purpurea (Sarraceniaceae),” American Journal of Botany, 89, 1024–1026.
Kleinn, C., and Vilčko, F. (2006), “Distance-Unbiased Estimation for Point-to-Tree Distance Sampling,” Canadian Journal of Forest Research, 36, 1407–1414.
Mackenzie, D. I., Nichols, J. D., Royle, J. A., Pollock, K. H., Bailey, L. L., and Hines, J. E. (2006), Occupancy Modeling and Estimation, San Diego: Academic Press.
Magnussen, S. (2012), “A New Composite k-Tree Estimator of Stem Density,” European Journal of Forest Research, 131, 1513–1527.
Magnussen, S., Fehrmann, L., and Platt, W. J. (2012), “An Adaptive Composite Density Estimator for k-Tree Sampling,” European Journal of Forest Research, 131, 307–320.
Murdoch, W. W. (1994), “Population Regulation in Theory and Practice,” Ecology, 75, 271–287.
Nothdurft, A., Saborowski, J., Nuske, R. S., and Stoyan, D. (2010), “Density Estimation Based on k-Tree Sampling and Point Pattern Reconstruction,” Canadian Journal of Forest Research, 40, 953–967.
Patil, S. A., Burnham, K. P., and Kovner, J. L. (1979), “Nonparametric Estimation of Plant Density by the Distance Method,” Biometrics, 35, 597–604.
Pyle, G. H., and Ehrlich, P. R. (2010), “Biological Collections and Ecological/environmental Research: a Review, Some Observations and a Look to the Future,” Biological Reviews, 85, 247–266.
Seber, G. A. F., and Salehi, M. M. (2012), Adaptive Sampling Designs: Inference for Sparse and Clustered Populations, New York: Springer.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ellison, A.M., Gotelli, N.J., Hsiang, N. et al. Kernel Intensity Estimation of 2-Dimensional Spatial Poisson Point Processes From k-Tree Sampling. JABES 19, 357–372 (2014). https://doi.org/10.1007/s13253-014-0175-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13253-014-0175-0