Abstract
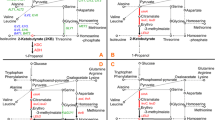
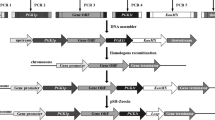
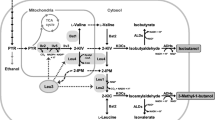
Pyruvate decarboxylase (PDC), a key enzyme in alcoholic fermentation in Saccharomyces cerevisiae, can degrade pyruvic acid to further convert acetaldehyde into ethanol. The main structural genes encoding PDC are PDC1 and PDC5. In this study, metabolic engineering principles were used to block the further metabolism of pyruvic acid; Saccharomyces cerevisiae Y2-1 with PDC1 disruption and Y2-15 with both PDC1 and PDC5 disruption were obtained using the LiAc/SS carrier DNA/PEG method. The specific PDC activity of mutant S. cerevisiae Y2-1 decreased by 31 % compared to that of the parent strain Y2, while specific PDC activity was barely detectable in mutant S. cerevisiae Y2-15. Moreover, the mutant Y2-1 with PDC1 disruption displayed no obvious effect on the rate of growth in the yeast nitrogen base with glucose (YNBG) medium, but the growth rate of S. cerevisiae Y2-15 was significantly lower than that of the parent strain Y2. Finally, through optimization of the fermentation medium, the accumulation of pyruvic acid by Y2-15 increased to 24.65 g/L over a period of 96 h, 16.86-fold higher than with the parental strain Y2 by shake flask cultivation.






Similar content being viewed by others
References
Barnett JA (1976) The utilization of sugars by yeasts. Adv Carbohydr Chem Biochem 32:125–234
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Burke D, Dawson D, Stearns T (2002) Methods in yeast genetics. Cold Spring Harbor Cold Spring Harbor Laboratory, New York
Candy JM, Duggleby RG, Mattick JS (1991) Expression of active yeast pyruvate decarboxylase in Escherichia coli. J Gen Microbiol 137:2811–2815
Causey TB, Shanmugam KT, Yomano LP, Ingram LO (2004) Engineering Escherichia coli efficient conversion of glucose to pyruvate. Proc Natl Acad Sci U S A 101:2235–2240
Ciriacy M, Breitenbach I (1979) Physiological effects of seven different blocks in glycolysis in Saccharomyces cerevisiae. J Bacteriol 139:152–160
Gao NF, Deng XH, Wang DP, Li L (2011) Determination of pyruvate decarboxylase activity in Saccharomyces cerevisiae. China Brewing 3:128–130 (In Chinese)
Gellissen G, Melber K, Janowicz ZA, Dahlems UM, Weydemann U, Piontek M, Strasser AWM, Hollenberg CP (1992) Heterologous protein production in yeast. Antonie Van Leeuwenhoek 62:79–93
Gietz RD, Schiestl RH (2007) Large-scale high-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc 2(1):38–41
Hohmann S (1991) Characterization of PDC6, a third structural gene for pyruvate decarboxylase in Saccharomyces cerevisiae. J Bacteriol 173(24):7963–7969
Hohmann S, Cederberg H (1990) Autoregulation may control the expression of yeast pyruvate decarboxylase structural genes PDC1 and PDC5. Eur J Biochem 188:615–621
Ishida N, Saitoh S, Onishi K, Tokuhiro T, Nagamori E, Kitamoto K, Takahashi H (2006) The effect of pyruvate decarboxylase gene knockout in Saccharomyces cerevisiae on L-lactic acid production. Biosci Biotechnol Biochem 70(5):1148–1153
Li Y, Hugenholtz J, Chen J, Lun SY (2002) Enhancement of pyruvate production by Torulopsis glabrata using a two-stage oxgen supply control strategy. Appl Microbiol Biotechnol 60:101–107
Liu Q, Yu L (2002) Yeast: A kind of model organism. Chem Life 20(2):61–65 (In Chinese)
Ostergaard S, Olsson L, Nielsen J (2002) Metabolic engineering of Saccharomyces cerevisiae. Microbiol Mol Biol Rev 64:34–50
Overkamp KM, Bakker BM, Kotter P, Luttik MA, van Dijken JP, Pronk JT (2002) Metabolic engineering of glycerol production in Saccharomyces cerevisiae. Appl Environ Microbiol 68:2814–2821
Porro D, Brambilla L, Ranzi BM, Martegani E, Alberghina L (1995) Development of metabolically engineered Saccharomyces cerevisiae cells for the production of lactic acid. Biotechnol Prog 11:294–298
Pronk JT, Steensma HY, van Dijken JP (1996) Pyruvate metabolism in Saccharomyces cerevisiae. Yeast 12:1607–1633
Sambrook JE, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Cold Spring Harbor Laboratory, New York
Schaaff I, Green JBA, Gozalbo D, Hohmann S (1989) A deletion of the PDC1 gene for pyruvate decarboxylase of yeast causes a different phenotype than previously isolated point mutations. Curr Genet 15:75–81
Schmitt HD, Zimmermann FK (1982) Genetic analysis of the pyruvate decarboxylase reaction in yeast glycolysis. J Bacteriol 151:1146–1152
Stockland AE, San Clemente CL (1969) Multiple forms of lactate dehydrogenase in Staphylococcus aureus. J Bacteriol 100:347–353
Tomar A, Eiteman MA, Altman E (2003) The effect of acetate pathway mutations on the production of pyruvate in Escherichia coli. Appl Microbiol Biotechnol 62:76–82
Van Maris AJA, Winkler AA, Porro D, van Dijken JP, Pronk JT (2004) Homofermentative lactate production cannot sustain annaerobic growth of engineered Saccharomyces cerevisiae: possible consequence of energy dependent lactate export. Appl Environ Microbiol 70:2898–2905
Wood V, Gwilliam R, Rajandream MA et al (2002) The genome sequence of Schizosaccharomyces pombe. Nature 415(6874):871–880
Zelić B, Gerharz T, Bott M, Vasić-Rački Đ, Wandrey C, Takors R (2003) Fed-batch process for pyruvate production by recombinant Escherichia coli YYC202 strain. Eng Life Sci 3:299–305
Zelić B, Gostović S, Vuorilehto K, Vasić-Rački Đ, Takors R (2004) Process strategies to enhance pyruvate production with recombinant Escherichia coli: from repetitive fed-batch to in situ product recovery with fully integrated electrodialysis. Biotechnol Bioeng 85:638–646
Acknowledgments
We thank Professor N.F. Gao for the valuable discussion. This work was supported by the National High-tech R&D Program of China (2012AA021302).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, D., Wang, L., Hou, L. et al. Metabolic engineering of Saccharomyces cerevisiae for accumulating pyruvic acid. Ann Microbiol 65, 2323–2331 (2015). https://doi.org/10.1007/s13213-015-1074-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13213-015-1074-5