Abstract
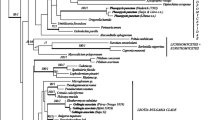
Graphis scripta, or script lichen, is a well-known species of crustose lichenized fungi, widely distributed in the temperate region of the Northern Hemisphere. It is now considered to be a species complex, but because of the lack of secondary chemistry and paucity of measurable morphological characters, species delimitation within the complex has been challenging and is thus far based on apothecium and ascospore morphology. In this study, we employed molecular as well as morphological data to assess phylogenetic structure and delimitation of lineages within the G. scripta complex. We generated sequences for four genetic markers (mtSSU, nuLSU, RPB2, and EF-1) and performed phylogenetic analyses. The resulting trees were used to determine the number of distinct lineages by applying a general mixed Yule-coalescent (GMYC) model and species tree estimation through maximum likelihood (STEM). Our analyses suggest between six and seven putative species within the G. scripta complex. However, these did not correspond to the taxa that were recently distinguished based on apothecium morphology and could not be circumscribed with the morphological characters that were traditionally used in the classification of the complex. Any formal taxonomic treatment will require additional sampling and evaluation of additional traits that potentially can characterize these clades.



Similar content being viewed by others
References
Acharius, E. (1809). Förteckning pa de i Sverige växande arter af Lafvarnes famille. Kongliga Vetenskaps Academiens Nya Handlingar, 30, 145–169.
Arup, U., & Åkelius, E. (2009). A taxonomic revision of Caloplaca herbidella and C. furfuracea. The Lichenologist, 41(5), 465–480.
Bickford, D., Lohman, D. J., Sodhi, N. S., Ng, P. K. L., Meier, R., Winker, K., et al. (2007). Cryptic species as a window on diversity and conservation. Trends in Ecology & Evolution, 22(3), 148–155.
Blanco, O., Crespo, A., Elix, J. A., Hawksworth, D. L., & Lumbsch, H. T. (2004). A molecular phylogeny and a new classification of parmelioid lichens containing Xanthoparmelia-type lichenan (Ascomycota: Lecanorales). Taxon, 53(4), 959–975.
Carstens, B. C., & Dewey, T. A. (2010). Species delimitation using a combined coalescent and information-theoretic approach: an example from North American myotis bats. Systematic Biology, 59(4), 400–414.
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution, 17, 540–552.
Coyne, J. A., & Orr, H. A. (2004). Speciation (pp. 1–545). Sinauer Associates Incorporated.
Cracraft, J. (1983). Species concepts and speciation analysis. In R. Johnston (Ed.), Current Ornithology (Vol. 1, pp. 159–187). Springer.
Crespo, A., & Lumbsch, H. T. (2010). Cryptic species in lichen-forming fungi. IMA Fungus, 1(2), 167–170.
Crespo, A., & Ortega, S. P. (2009). Cryptic species and species pairs in lichens: a discussion on the relationship between molecular. Anales del Jardin Botánico de Madrid, 66(1), 71–81.
Czarnota, P., & Guzow-Krzemińska, B. (2010). A phylogenetic study of the Micarea prasina group shows that Micarea micrococca includes three distinct lineages. The Lichenologist, 42(1), 7–21.
Darriba, D., Taboada, G. L., Doallo, R., & Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nature Methods, 9(8), 772.
de Queiroz, K. (2007). Species concepts and species delimitation. Systematic Biology, 56(6), 879–886.
Divakar, P. K., & Crespo, A. (2015). Molecular phylogenetic and phylogenomic approaches in studies of lichen systematics and evolution. In Recent Advances in Lichenology (pp. 45–60). New Delhi: Springer.
Drummond, A. J., Ashton, B., Buxton, S., Cheung, M., Cooper, A., Duran, C., et al. (2014). Geneious v. 8.0.3. Biomatters.
Erichsen, C. F. E. (1957). Flechtenflora von Nordwestdeutschland (pp. 1–411). Stuttgart: Gustav Fischer.
Ezard, T., Fujisawa, T., & Barraclough, T. G. (2009). SPLITS: SPecies’ LImits by Threshold Statistics. Retrieved from http://R-Forge.R-project.org/projects/splits/
Fontaneto, D., Iakovenko, N., Eyres, I., Kaya, M., Wyman, M., & Barraclough, T. G. (2011). Cryptic diversity in the genus Adineta Hudson & Gosse, 1886 (Rotifera: Bdelloidea: Adinetidae): a DNA taxonomy approach. Hydrobiologia, 662(1), 27–33.
Fujisawa, T., & Barraclough, T. G. (2013). Delimiting species using single-locus data and the generalized mixed Yule coalescent approach: a revised method and evaluation on simulated data sets. Systematic Biology, 62(5), 707–724.
Garland, T., Jr., Dickerman, A. W., Janis, C. M., & Jones, J. A. (1993). Phylogenetic analysis of covariance by computer simulation. Systematic Biology, 42(3), 265–292.
Goldstein, P. Z., DeSalle, R., Amato, G., & Vogler, A. P. (2000). Conservation genetics at the species boundary. Conservation Biology, 14(1), 120–131.
Guindon, S., & Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology, 52(5), 696–704.
Hodkinson, B. P., & Lendemer, J. C. (2011). Molecular analyses reveal semi-cryptic species in Xanthoparmelia tasmanica. Bibliotheca Lichenologica, 106, 108–119.
Högnabba, F., & Wedin, M. (2003). Molecular phylogeny of the Sphaerophorus globosus species complex. Cladistics, 19(3), 224–232.
Hudson, R. R. (1990). Gene genealogies and the coalescent process. In D. J. Futuyma & J. Antonivics (Eds.), Oxford surveys in evolutionary biology (Vol. 7, pp. 1–44). Oxford: Oxford University Press.
Huelsenbeck, J. P., & Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics (Oxford, England), 17, 754–755.
Kraichak, E., Parnmen, S., Lücking, R., Rivas Plata, E., Aptroot, A., Cáceres, M. E. S., et al. (2014). Revisiting the phylogeny of Ocellularieae, the second largest tribe within Graphidaceae (lichenized Ascomycota: Ostropales). Phytotaxa, 189, 52–81.
Kubatko, L. S., Carstens, B. C., & Knowles, L. L. (2009). STEM: species tree estimation using maximum likelihood for gene trees under coalescence. Bioinformatics (Oxford, England), 25(7), 971–973.
Leavitt, S. D. S., Fankhauser, J. D. J., Leavitt, D. H. D., Porter, L. D. L., Johnson, L. A. L., & Clair, L. L. L. S. (2011). Complex patterns of speciation in cosmopolitan “rock posy” lichens—discovering and delimiting cryptic fungal species in the lichen-forming Rhizoplaca melanophthalma species-complex (Lecanoraceae, Ascomycota). Molecular Phylogenetics and Evolution, 59(3), 587–602.
Leavitt, S. D., Moreau, C. S., & Lumbsch, H. T. (2015). The dynamic discipline of species delimitation: progress toward effectively recognizing species boundaries in natural populations. In Recent Advances in Lichenology (pp. 11–44). New Delhi: Springer.
Lendemer, J. C. J. (2011). A taxonomic revision of the North American species of Lepraria s.l. that produce divaricatic acid, with notes on the type species of the genus L. incana. Mycologia, 103(6), 1216–1229.
Lendemer, J. C., & Hodkinson, B. P. (2013). A radical shift in the taxonomy of Lepraria s.l.: molecular and morphological studies shed new light on the evolution of asexuality and lichen growth form diversification. Mycologia, 105(4), 994–1018.
Linnaeus, C. (1753). Species plantarum (pp. 1–639). Stockholm: Salvius.
Lücking, R., Archer, A. W., & Aptroot, A. (2009). A world-wide key to the genus Graphis (Ostropales: Graphidaceae). The Lichenologist, 41(04), 363–452.
Lücking, R., Dal-Forno, M., Lawrey, J. D., Bungartz, F., Rojas, M. E. H., Hernández, J. E., et al. (2013). Ten new species of lichenized Basidiomycota in the genera Dictyonema and Cora (Agaricales: Hygrophoraceae), with a key to all accepted genera and species in the Dictyonema clade. Phytotaxa, 139(1), 1–38.
Lücking, R., Johnston, M. K., Aptroot, A., Kraichak, E., Lendemer, J. C., Boonpragob, K., et al. (2014). One hundred and seventy five new species of Graphidaceae: closing the gap or a drop in the bucket? Phytotaxa, 189, 7–38.
Lumbsch, H. T., & Leavitt, S. D. (2011). Goodbye morphology? A paradigm shift in the delimitation of species in lichenized fungi. Fungal Diversity, 50(1), 59–72.
Lumbsch, H. T., Mangold, A., Martin, M. P., & Elix, J. A. (2008). Species recognition and phylogeny of Thelotrema species in Australia (Ostropales, Ascomycota). Australian Systematic Botany, 21(3), 217–227.
Lumbsch, H. T., Parnmen, S., Rangsiruji, A., & Elix, J. A. (2010). Phenotypic disparity and adaptive radiation in the genus Cladia (Lecanorales, Ascomycota). Australian Systematic Botany, 23(4), 239–247.
Lumbsch, H. T., Parnmen, S., Kraichak, E., Papong, K. B., & Lücking, R. (2014). High frequency of character transformations is phylogenetically structured within the lichenized fungal family Graphidaceae (Ascomycota: Ostropales). Systematics and Biodiversity, 12(3), 271–291.
Mangold, A., Martin, M. P., Lücking, R., & Lumbsch, H. T. (2008). Molecular phylogeny suggests synonymy of Thelotremataceae within Graphidaceae (Ascomycota : Ostropales). Taxon, 57, 476–486.
Mayr, E. (1963). Animal species and evolution (pp. 1–797). Cambridge: Harvard University Press.
McCune, B., & Schoch, C. (2009). Hypogymnia minilobata (Parmeliaceae), a New Lichen from Coastal California. The Bryologist, 112(1), 94–100.
Miller, M. A., Pfeiffer, W., & Schwartz, T. (2010). Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop (GCE), 2010, 1–8.
Moncada, B., Coca, L. F., & Lücking, R. (2013). Neotropical members of Sticta (lichenized Ascomycota: Lobariaceae) forming photosymbiodemes, with the description of seven new species. The Bryologist, 116(2), 169–200.
Nee, S., May, R. M., & Harvey, P. H. (1994). The reconstructed evolutionary process. Philosophical Transactions: Biological Sciences, 344(1309), 305–311.
Neuwirth, G., & Aptroot, A. (2011). Recognition of four morphologically distinct species in the Graphis scripta complex in Europe. Herzogia, 24, 207–230.
Paradis, E. (2010). pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics (Oxford, England), 26, 419–420.
Paradis, E. (2013). Molecular dating of phylogenies by likelihood methods: a comparison of models and a new information criterion. Molecular Phylogenetics and Evolution, 67(2), 436–444. doi:10.1016/j.ympev.2013.02.008.
Paradis, E., Claude, J., & Strimmer, K. (2004). APE: analyses of phylogeneticsand evolution in R language. Bioinformatics, 20, 289–290.
Parnmen, S., Rangsiruji, A., Mongkolsuk, P., Boonpragob, K., Elix, J. A., & Lumbsch, H. T. (2010). Morphological disparity in Cladoniaceae: the foliose genus Heterodea evolved from fruticose Cladia species (Lecanorales, lichenized Ascomycota). Taxon, 59(3), 841–849.
Parnmen, S., Rangsiruji, A., Mongkolsuk, P., Boonpragob, K., Nutakki, A., & Lumbsch, H. T. (2012). Using phylogenetic and coalescent methods to understand the species diversity in the Cladia aggregata complex (Ascomycota, Lecanorales). PLoS ONE, 7(12), e52245.
Parnmen, S., Cáceres, M. E. S., Lücking, R., & Lumbsch, H. T. (2013). Myriochapsa and Nitidochapsa, two new genera in Graphidaceae (Ascomycota: Ostropales) for chroodiscoid species in the Ocellularia clade. The Bryologist, 116(2), 127–133.
Pillon, Y., Hopkins, H. C. F., Rigault, F., Jaffré, T., & Stacy, E. A. (2014). Cryptic adaptive radiation in tropical forest trees in New Caledonia. New Phytologist, 202(2), 521–530.
Pons, J., Barraclough, T., Gomez-Zurita, J., Cardoso, A., Duran, D., Hazell, S., et al. (2006). Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Systematic Biology, 55(4), 595–609.
Printzen, C., & Ekman, S. (2002). Genetic variability and its geographical distribution in the widely disjunct Cavernularia hultenii. The Lichenologist, 34(2), 101–111.
Printzen, C., Ekman, S., & Tønsberg, T. (2003). Phylogeography of Cavernularia hultenii: evidence of slow genetic drift in a widely disjunct lichen. Molecular Ecology, 12(6), 1473–1486.
Rambaut, A. (2012). FigTree. Version 1.4.2.
Rehner, S. (2001). Primers for Elongation Factor 1-α (EF1-α). Assembling the fungal tree of life. Retrieved from http://www.aftol.org/pdfs/EF1primer.pdf
Revell, L. J. (2011). phytools: an R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution, 3, 217–223.
Rivas Plata, E., & Lumbsch, H. T. (2011). Parallel evolution and phenotypic divergence in lichenized fungi: a case study in the lichen-forming fungal family Graphidaceae (Ascomycota: Lecanoromycetes: Ostropales). Molecular Phylogenetics and Evolution, 61(1), 45–63.
Rivas Plata, E., Lücking, R., & Lumbsch, H. T. (2012). A new classification for the lichen family Graphidaceae s.lat. (Ascomycota: Lecanoromycetes: Ostropales). Fungal Diversity, 52, 107–121.
Rivas Plata, E., Parnmen, S., Staiger, B., Mangold, A., Frisch, A., Weerakoon, G., Hernandez, J., et al. (2013). A molecular phylogeny of Graphidaceae (Ascomycota, Lecanoromycetes, Ostropales) including 428 species. MycoKeys, 6, 55–94.
Ruprecht, U., Lumbsch, H. T., Brunauer, G., Green, T. A., & Türk, R. (2010). Diversity of Lecidea (Lecideaceae, Ascomycota) species revealed by molecular data and morphological characters. Antarctic Science, 22(6), 727.
Sanderson, M. J. (2002). Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Molecular Biology and Evolution, 19(1), 101–109.
Schneider, C. A., Rasband, W. S., & Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nature Methods, 9, 671–675.
Spribille, T., Klug, B., & Mayrhofer, H. (2011). A phylogenetic analysis of the boreal lichen Mycoblastus sanguinarius (Mycoblastaceae, lichenized Ascomycota) reveals cryptic clades correlated with fatty acid profiles. Molecular Phylogenetics and Evolution, 59(3), 603–614.
Staiger, B. (2002). Die Flechtenfamilie Graphidaceae. Stuttgart: Schweizerbart’sche Verlagsbuchhandlung.
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics (Oxford, England), 22, 2688–2690.
Stamatakis, A., Hoover, P., & Rougemont, J. (2008). A Rapid Bootstrap Algorithm for the RAxML Web Servers. Systematic Biology, 57, 758–771.
Stenroos, S. K., & DePriest, P. T. (1998). SSU rDNA phylogeny of cladoniiform lichens. American Journal of Botany, 85(11), 1548–1559.
Talavera, G., & Castresana, J. (2007). Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Systematic Biology, 56, 564–577.
Tehler, A., & Irestedt, M. (2007). Parallel evolution of lichen growth forms in the family Roccellaceae (Arthoniales, Ascomycota). Cladistics, 23(5), 432–454.
Vilgalys, R., & Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology, 172, 4238–4246.
Vogler, A. P., & Monaghan, M. T. (2007). Recent advances in DNA taxonomy. Journal of Zoological Systematics and Evolutionary Research, 45(1), 1–10.
Vondrák, J. (2012). Biomonitoring, ecology, and systematics of lichens. The Bryologist, 115(4), 636–637.
Vondrák, J., Říha, P., Arup, U., & Søchting, U. (2009). The taxonomy of the Caloplaca citrina group (Teloschistaceae) in the Black Sea region; with contributions to the cryptic species concept in lichenology. The Lichenologist, 41(6), 571–604.
Wedin, M., & Döring, H. (1999). The phylogenetic relationship of the Sphaerophoraceae, Austropeltum and Neophyllis (lichenized Ascomycota) inferred by SSU rDNA sequences. Mycological Research, 103(9), 1131–1137.
Wedin, M., Westberg, M., Crewe, A. T., & Tehler, A. (2009). Species delimitation and evolution of metal bioaccumulation in the lichenized Acarospora smaragdula (Ascomycota, Fungi) complex. Cladistics, 25(2), 161–172.
Wiens, J. J., & Penkrot, T. A. (2002). Delimiting species using DNA and morphological variation and discordant species limits in spiny lizards (Sceloporus). Systematic Biology, 51(1), 69–91.
Wirtz, N., Printzen, C., & Lumbsch, H. T. (2008). The delimitation of Antarctic and bipolar species of neuropogonoid Usnea (Ascomycota, Lecanorales): a cohesion approach of species recognition for the Usnea perpusilla complex. Mycological Research, 112, 472–484.
Yost, J. M., Barry, T., Kay, K. M., & Rajakaruna, N. (2012). Edaphic adaptation maintains the coexistence of two cryptic species on serpentine soils. American Journal of Botany, 99(5), 890–897.
Zahlbruckner, A. (1923). Catalogus lichenum universalis (Vol. 2, pp. 1–701). Leipzig: Gebrüder Borntraeger.
Zhang, J., Kapli, P., Pavlidis, P., & Stamatakis, A. (2013). A general species delimitation method with applications to phylogenetic placements. Bioinformatics (Oxford, England), 29(22), 2869–2876.
Zoller, S., Scheidegger, C., & Sperisen, C. (1999). PCR primers for the amplification of mitochondrial small subunit ribosomal DNA of lichen-forming ascomycetes. The Lichenologist, 31, 511–516.
Acknowledgments
We would like to thank Kyle Huff (Roosevelt University) for helping with morphological measurements. The molecular work was performed at the Pritzker Laboratory for Molecular Systematics and Evolution at The Field Museum (Chicago).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kraichak, E., Lücking, R., Aptroot, A. et al. Hidden diversity in the morphologically variable script lichen (Graphis scripta) complex (Ascomycota, Ostropales, Graphidaceae). Org Divers Evol 15, 447–458 (2015). https://doi.org/10.1007/s13127-015-0219-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13127-015-0219-5