Abstract
Primary breast carcinomas expressing both estrogen and progesterone receptors are most likely to respond to tamoxifen therapy, especially in patients with early-stage lesions. However, certain patients exhibit clinicopathologic features suggesting good prognosis relapse within 10 years, justifying a search for biomarkers identifying patients at risk for recurrence. Nine candidate genes associated with estrogen signaling were selected from microarray studies and combined with those for conventional biomarkers (ESR1, PGR, ERBB2). Expression of this 12-gene subset was analyzed by RT-qPCR in frozen tissue specimens from 60 early-stage, estrogen receptor (ER)+/progestin receptor (PR)+ breast cancers from patients treated with adjuvant tamoxifen. A multivariate model was created by Cox regression using a training data set and applied to an independent validation set. A five-gene model was developed from the training set (n = 36) that exhibited significant correlations with both relapse-free and overall survival. Applying this model to Kaplan–Meier regression, patients were separated into low-risk (100% relapse-free at 150 months) and high-risk (60% relapse-free at 150 months) groups (P = 0.03). When this model was applied to the validation set (n = 24), similar risk stratification was achieved for both relapse-free and overall survival (P = 0.01 and 0.04, respectively). We developed a five-gene model composed of PgR, BCL2, ERBB4 JM-a, RERG, and CD34 that identified early-stage, ER+/PR+ breast cancers in patients treated with tamoxifen that relapsed, although they exhibited clinicopathologic features suggesting good prognosis. Within this multivariate model, increased expression of PgR, ERBB4 JM-a, RERG, and CD34 was associated with increased survival, while increased expression of BCL2 was associated with decreased survival.
Similar content being viewed by others
Introduction
Recognition of the role of estrogen and estrogen receptor (ER) signaling in breast carcinoma development and progression led to development of targeted endocrine therapies for hormone-dependent breast cancers (e.g., [1]). Tamoxifen, a widely used anti-estrogen therapy for primary breast cancer, is classified as a selective ER modulator (SERM) which acts by competitive antagonism of the cognate ligand for the ER, resulting in inhibition of growth and proliferation. Estrogen (ER) and progestin receptors (PR) are biomarkers for breast cancer assessment of response to anti-estrogen therapy (e.g., [2–5]). Currently, patients treated with tamoxifen exhibit a response rate of 50–80% for breast cancers positive for ER and/or PR compared to only 5–10% response for cancers not expressing detectable levels of either receptor protein [6, 7].
Increased expression of ER and PR proteins in human breast cancers is also a weak predictor of disease-free and overall survival (OS; e.g., [3–5, 8, 9]). In general, patients with ER+/PR+ breast cancers exhibit better prognosis than those with ER−/PR− status. Patients with ER+/PR− and ER−/PR+ tumors usually exhibit intermediate prognosis with more significant differences observed in later-stage breast cancers [10].
Although ER status is the strongest predictor of tamoxifen response, PR status independently predicts tamoxifen response in both ER-positive [11] and ER-negative [7] cancers. Patients with ER+/PR+ breast cancers are reported to exhibit higher response rates than those with carcinomas positive for only one of the receptors (e.g., [11–13]). For example, Bardou et al. revealed significant differences in time to recurrence and mortality as a function of ER/PR status in patients treated with adjuvant tamoxifen, using data from 1,688 patients [12]. Compared to ER−/PR− cancers, relative risk reduction for recurrence was 25% and 53% for ER+/PR− and ER+/PR+ cancers, respectively. It is evident ER status alone predicts prognosis and response to endocrine therapy with less efficacy than combined expression of ER and PR, indicating ER+/PR+ breast cancers represent a population most likely to respond to tamoxifen therapy. However, 20–25% of tamoxifen-treated patients within the ER+/PR+ cancer population exhibit a recurrence within 10 years [1, 14, 15].
Commercial gene expression assays in the USA are available, with the most widely used being OncotypeDx™ (Genomic Health, Inc.), designed to determine risk of recurrence in tamoxifen-treated patients with ER-positive breast cancers. OncotypeDx™ is a qPCR-based assay that determines likelihood of recurrence (recurrence score) using a 21-gene panel, containing 16 cancer-related genes and five reference genes [16, 17]. For patients treated with adjuvant tamoxifen, this assay significantly predicted those at higher risk of recurrence [16]. While candidates for this assay must have ER-positive breast cancers, there was no regard for PR status in the validation studies. PR also was not included in the molecular signature reported by Vendrell et al. for predicting tamoxifen failure [18]. This represents a potential drawback considering the importance for PR status for both prognosis and tamoxifen response in ER-positive breast cancers [7, 11, 12].
Our laboratory and Arcturus Applied Genomics (now Molecular Devices) incorporated use of laser capture microdissection (LCM) to procure pure populations of breast carcinoma cells for microarray analyses of gene expression [19, 20]. Global gene expression profiles, analyzed by hierarchical clustering, revealed 200 over-expressed genes distinguishing four molecular subtypes, two of which were associated predominantly with ER-positive breast carcinomas exhibiting distinct survival characteristics [19]. The set of 200 candidate genes were selected from our earlier studies of LCM-procured carcinoma cells since these cells are the target for therapeutic manipulations. However, the use of LCM collected specific cell types only served as a discovery tool for our long-term goal to define clinically relevant gene subsets, avoiding the use of LCM in a routine gene expression test.
Since the focus of our study was hormone-dependent (i.e., ER+/PR+) breast cancers treated with tamoxifen which affects ERE-dependent gene transcription, the list of genes within the two ER-positive subtypes was examined for evidence of both estrogen-dependent and ERE-dependent expression. Using the web-based program ERTargetDB [21] and additional literature review, we identified 16 genes from the two ER-positive subtypes that also exhibited estrogen-responsive gene expression. Additionally, there were 13 genes identified within these two ER-positive subtypes of breast cancer that exhibited known or candidate ERE sequences. Nine genes, which appeared in each of the two lists, were selected for qPCR analyses using total RNA from frozen breast cancer biopsies. Expression of these genes was examined with that of ESR1, PGR, and ERBB2 genes for conventional breast cancer biomarkers, ER, PR, and HER2, respectively. While LCM is a useful tool for global gene expression profiling using microarray, it is impractical for use in a routine clinical test of gene expression. Furthermore, techniques such as qPCR, which focus on specific analytes, easily provide clinically relevant information using intact tissue. For the purposes of this study, sections of intact tissue specimens exhibiting significant cancer cell content were used for the qPCR analyses reported.
The population studied was composed of patients with stages 1–2B cancers without distant metastasis present at diagnosis, excluding those with advanced stage cancers. The early-stage, ER+/PR+ breast carcinomas selected represent a population that generally is considered to have good prognosis [3, 4, 9, 22]. Using training (n = 36) and validation (n = 24) populations, we identified a subset of five estrogen-regulated genes in ER+/PR+, early-stage breast cancers treated with adjuvant tamoxifen which classify patients with high risk of recurrence that are uncharacteristic of their clinicopathologic status.
Methods and Materials
Success of this investigation requires an accurate selection of breast carcinomas in which levels of ER and PR were quantified according to clinically accepted criteria (e.g., [4, 23, 24]). Furthermore, genomic analyses critical to our goal must be performed on frozen tissue biopsies processed under stringent conditions required in genomic studies estimating labile macromolecules (e.g., [16, 17, 20]).
Specimen Selection
All tissue specimens and follow-up information were de-identified and encoded in the Tumor Marker database established by our laboratory, and no identifiers were available in any part of this research according to Institutional Review Board policies. Primary breast carcinoma tissue specimens, collected between 1988 and 1997, were selected with associated ER/PR protein levels, quantified by radio-ligand binding or enzyme immunoassays under highly controlled conditions [2, 4, 23, 24]. All assays were performed within the clinically licensed (CLIA-approved) Hormone Receptor Laboratory, using FDA-approved reagents and protocols. For study specimens, clinicopathologic and longitudinal data were collected.
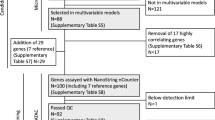
A qPCR dataset was created using tissue specimens with associated clinical follow-up from 279 breast cancer patients in order to determine the clinical relevance of expression of the 12 gene subset. Within this population, there were 60 female patients meeting the criteria (i.e., ER+/PR+, stages 1–2 breast cancers, treated with adjuvant tamoxifen) with frozen tissue available that passed quality assessment standards described below. This study population (Table 1) was selected with adherence to REMARK guidelines [25]. For the patients classified as relapse-free, only one patient had <50 months of follow-up (31 months). Modified radical mastectomy was the surgical therapy chosen for 56 cases, and partial mastectomy (lumpectomy) was used for four cases with external beam radiation therapy. All patients received adjuvant tamoxifen therapy, although eight patients did receive additional chemotherapy (Table 1). The analysis shown in Fig. 3 was obtained from the entire dataset of 279 cases. This population includes women with both ER-negative and late-stage breast cancers.
Tissue Preparation
All tissue preparation was performed in a nuclease/protease-free environment. Each de-identified tissue specimen was collected according to stringent policies to preserve both structural and molecular integrity [2, 4]. Residual tissue was stored at −80°C in sterile, cryogenic vials prior to processing on dry ice, embedding in cassettes containing Tissue Tek® O.C.T.® compound (VWR Scientific, West Chester, PA, USA) and immediately freezing in a dry ice/isopentane bath for 30–60 s [20]. For RNA extraction, three to five 20-μm tissue sections were placed into pre-chilled 1.5 ml microcentrifuge tubes then stored on dry ice. For histology, 7-μm sections were prepared on room-temperature charged microscope slides and stored at −80°C until further processing.
Hematoxylin and Eosin Staining
Slides were transferred directly from −80°C to alcoholic formalin (nine parts 70% ethanol in water, one part formaldehyde) for 5 min, then transferred to dH2O for 4 min with slight agitation, followed by hematoxylin I (Fisher Scientific, Pittsburgh, PA, USA) for 3 min and running water for 3 min. Slides were dipped in ammonium, rinsed in water, then dipped in Eosin Y (Thermo Shandon, Pittsburgh, PA, USA). Slides were dehydrated in 95% ethanol for 2 min, 100% ethanol for 3 min, and then xylenes for 6 min. After drying for 10 min, a glass coverslip was applied with Permount (Fisher Scientific). Slides were examined by light microscopy, and cellular content (cancer cells, inflammatory cells, stromal cells) was recorded for each specimen. Specimens selected contained ≥40% carcinoma cells and ≤10% inflammatory cells (predominantly lymphocytes).
RNA Isolation
Total RNA was isolated by spin-column chromatography using RNeasy™ (Qiagen, Valencia, CA, USA) reagents. Tissue sections were mixed vigorously for 1 min in 350 μl RLT buffer (1% β-mercaptoethanol), frozen on dry ice, then thawed, and mixed again for 1 min. Column loading, washing, and RNA elution were performed as per manufacturer’s protocol. Total RNA was analyzed by electrophoresis using a Bioanalyzer™ (Agilent Technologies, Santa Clara, CA, USA). RNA integrity was determined from the 18S/28S ratio and the RNA integrity number (RIN) [26]. rRNA ratios of 1.5–2.0 and RIN values ≥7 generally indicate intact RNA. Specimens with poor quality RNA were excluded from this study.
Reverse Transcription
Total RNA (100–500 ng in 10 μl) was combined with 166 ng random hexamers (Promega, Madison, WI, USA), 10 nmol of dNTPs (Invitrogen, Carlsbad, CA, USA), and nuclease-free H2O for a total volume of 13 μl. After allowing primers to anneal for 5 min at 65°C, an enzyme mix containing 1 μl SuperScript™ III reverse transcriptase, 4 μl 2× Master Mix (Invitrogen), 1 μl RNAsin™ RNAse inhibitor (Promega), and 1 μl dithiothreitol was added. Each reaction was incubated at 25°C for 5 min, followed by 55°C for 60 min, then 75°C for 15 min to prepare the cDNA, which was stored at −20°C.
qPCR Analyses
Real-time PCR was performed using SYBR™ Green detection (Applied Biosystems, Carlsbad, CA, USA). cDNA was diluted 20-fold in 2 ng/μl polyinositol (Sigma, St. Louis, MO, USA) and combined with 2× SYBR™ Green master mix for each 12 μl reaction containing forward and reverse primers (300 nM final concentration). PCR was performed using 40 cycles (58°C for 30 s, 72°C for 30 s, then 95°C for 15 s). For each experiment, reactions were completed in duplicate wells using the 7900HT Sequence Detection System (Applied Biosystems). Using β-actin as the reference gene (forward primer ATCCCCCAAAGTTCACAATG, reverse primer GTGGCTTTTAGGATGGCAAG), relative expression levels of each target gene were determined by the ΔΔCt method [27]. Human Universal Reference RNA (Stratagene, Santa Clara, CA, USA) was analyzed by qPCR in parallel (500 ng per reverse transcription reaction) as a calibrator in ΔΔCt calculations. Experiments were performed in triplicate to calculate a mean ± standard deviation. Both no template and RNA (no RT) reactions were used as negative controls.
Primers were designed using each gene sequence as a template for the Primer Express™ (Applied Biosystems) PCR primer selection tool. Based on previous observations suggesting a clinical role for the protein product of the ERBB4 JM-a splice variant (e.g., [28]), we developed primers to measure specific expression of this splice variant. Sequences used for each of the 13 genes were as follows: ESR1 (forward—CCAAATTGTGTTTGATGGATTAA, reverse—GACAAAACCGAGTCACATCAGTAATAG), PGR (forward—TCCTGGGAATGGGCTGTAC, reverse—AAACTCGTGCATGCTGTGAAG), ERBB2 (forward—AAAAGCGACCCATTCAGAGACT, reverse—AAAAACTAAACAGAAAAGCACTCTGTACAA), BCL2 (forward—GCCCCAAAAGGAGAAGAACATC, reverse—TTCTGCCCCTGCCAAATCT), CAXII (forward—CAGGCGCAACTCCTCCATT, reverse—GGTCGGTTCCTTCTCAGTCATG), ERBB4 JM-a (forward—GGCCATTCCACTTTACCACAA, reverse—CAGAATGAAGAGCCCACCAATT), RERG (forward—CTCCAGGCAGGTTAGCACAGA, reverse primer AGGCAGAGCACTCGTAAAAAGC), CD34 (forward—CTCCAGAAACGGCCATTCAG, reverse—CCCACCTAGCCGAGTCA CAA), EDG1 (forward—CTCTTCTGCACCACGGTCTTC, reverse—CTCCGAGTCCTGACCAAGGA), NQO1 (forward—GATTGGACCGAGCTGGAAAAC, reverse—CAGCCGTCAGCTATTGTGGATA), PTGDS (forward—AAATTCACCGCCTTCTGCAA, reverse—TGTTCCGTCATGCACTTATCG), and SDF1 (forward—GGGAAATATTCCCTAGAAACTTCCA, reverse—GAGTCCAGCGAGGTTGCAA).
Statistical Analyses
All qPCR data were log2-transformed before downstream analyses. For all analyses, a P value of ≤0.05 was accepted as statistically significant. Variance analyses (e.g., chi-squared, Mann–Whitney, and Kruskal–Wallis), Cox regression, and graphical analyses (e.g., scatter plot, receiver operating characteristic (ROC), and Kaplan–Meier regression) were performed using PASW Statistics 18.0 (SPSS, Inc., Chicago, IL, USA) and GraphPad Prism™ 4 (GraphPad, Inc., La Jolla, CA, USA). Cox regression analyses were performed using both forward and backward conditional selection methods. The multi-gene models obtained by Cox regression were applied to the following equation [29] to create an odds value for each case: Odds = e^(B 1 X 1 + B 2 X 2 + ….+B n X n ), where B (beta) is a coefficient calculated for each gene using the Cox proportional hazards model and X is the log2-transformed value of relative gene expression determine by qPCR. Negative B coefficients indicate that decreased expression is associated with decreased survival, which also corresponds to hazard ratios <1. Lower odds values, which are calculated for each patient, are associated with decreased risk of recurrence/mortality. Cases were grouped according to increasing odds values for categorical analyses using Kaplan–Meier regression.
Results
A Five-Gene Model Was Generated Using qPCR Results from the 12-Gene Subset
The study population was sorted randomly, then divided into a training set (n = 36) and a validation set (n = 24). Significance of expression of each of the 12 genes relative to relapse-free survival (RFS) was determined by univariate Cox regression relating qPCR results to clinical outcome in the training set. None of the 12 candidate genes exhibited independent associations with RFS in the study population (P > 0.05). Multivariate Cox regression was performed using both forward conditional and backward conditional selection [29]. Since expression of each gene did not exhibit an independent association with RFS, forward selection did not reveal any clinically relevant candidates. However, backward selection revealed five genes, whose collective expression was associated with RFS (Table 2). Expression of four genes (PgR, ERBB4 JM-a, RERG, and CD34) were positively associated with RFS (hazard ratios <1.00). Expression of BCL2, however, exhibited a negative impact on prediction of RFS in this model. A higher absolute value of the B coefficient gives more weight to that variable, making CD34 and BCL2 the most influential variables contributing to the overall odds value for each case.
Prognostic Model is Associated with RFS and OS in the Training Population
The B coefficients determined from the five-gene model (Table 2) were applied to the equation described in “Methods and Materials” section to obtain an odds value for each case. Odds values were compared to recurrence status of the training population using ROC analysis (Fig. 1a) which calculates sensitivity and 1-specificity for predicting relapse using each of the 36 data points as a potential cutoff. This analysis provides an overall indication of the power of the model for predicting relapse, as shown by an area under the curve (AUC) value approaching 1 (Fig. 1a). Similar ROC analysis was performed for OS, in which the AUC was 0.88 (95% confidence interval (CI) 0.77–0.99, P = 0.01) indicating a similar power for predicting OS. Mortality is defined as death due to breast carcinoma.
Prognostic significance of the five-gene model in the training population. ROC analysis was utilized to compare the five-gene model with RFS (a) in the training set (n = 36). The AUC for RFS was 0.87 (95% CI 0.75–0.99, P = 0.01). Patients were categorized on the basis of their risk scores into low-, intermediate-, and high-risk groups (n = 12 for each group) and RFS for each group compared using Kaplan–Meier curves (b). Logrank test indicated significant separation among groups (P = 0.03) with a significant trend (P = 0.01). Intermediate- and high-risk groups were combined for Kaplan–Meier analyses of RFS (c) and OS (d). Logrank test indicated significant differences in RFS and OS for the low- and high-risk groups (P = 0.03 and 0.04, respectively)
Odds values obtained for cases in the training population were also used for categorical comparisons by Kaplan–Meier regression. Data sets were initially divided into thirds, representing low-, intermediate-, and high-risk groups with increasing odds values in each group, as suggested by Paik et al. [16] and Sparano and Paik [17]. There was a progressive increase in incidence of breast cancer recurrence for intermediate- and high-risk groups (Fig. 1b) with the low-risk group remaining recurrence-free at 150 months. The odds value cutoff separating low- and intermediate-risk groups was also applied to the validation set (described below), and the intermediate- and high-risk groups were combined for comparison with the low-risk group (Fig. 1c, d). There was significant separation between clinical outcome of the two groups for RFS and OS. The low-risk group remained recurrence-free at 150 months, while approximately 55% of breast cancer patients in the intermediate-/high-risk group remained recurrence-free at 150 months. The hazard ratios could not be determined because there were no events in the low-risk group; however, the logrank test was significant for both groups (chi-squared of 4.93 and 4.11, respectively).
The Five-Gene Model Predicts RFS and OS in the Validation Set
qPCR results from breast carcinoma specimens in the validation set were applied to the equation described in “Methods and Materials” section to obtain odds values for each specimen. Odds values were then compared to clinical outcome using both ROC and Kaplan–Meier analyses (Fig. 2). ROC curve AUC values for both relapse and mortality (Fig. 2a, b) were comparable to those obtained in the training set (Fig. 1a). For categorical analysis of survival probabilities, the same odds value cutoff that separated low- and intermediate-risk groups in the training set (6.6E−8) was applied to the validation set. This cutoff value divided the validation set into low- and high-risk groups (Fig. 2c, d). Although patients in the low-risk group did not exhibit any relapse or mortality after 150 months of follow-up, approximately 50% of patients in the high-risk group remained recurrence-free at 150 months.
Prognostic significance of the five-gene model in the validation population. ROC analyses were utilized to compare the five-gene model with RFS (a) and OS (b) in the validation set (n = 24). The AUC for RFS was 0.82 (95% CI 0.65–0.99, P = 0.03). The AUC for OS was 0.82 (95% CI 0.65–0.99, P = 0.03). Patients were categorized by their risk scores into low- and high-risk groups (n = 12 for each group). RFS (c) and OS (d) for each group were compared using Kaplan–Meier curves. Logrank test indicated significant differences in both RFS and OS of patients in low- and high-risk groups (P = 0.01 and 0.04, respectively)
There Are No Significant Differences in the Clinical Parameters of Cases in the Low- and High-Risk Groups Assigned to Either the Training or Validation Sets
Gene expression results from training and validation sets provided significant separation of patients with different clinical outcomes using the same odds value cutoff. For each population, clinical parameters were compared between low- and high-risk patient groups (Table 3). Intermediate- and high-risk groups were combined in the training set since this odds value cutoff was also applied to the validation set. For grade, nodal status, and additional therapies, chi-squared analysis was used to compare groups. Age and tumor size were compared between groups using the Mann–Whitney test. There were no significant differences between risk groups regarding age, nodal status, grade, tumor size, or additional chemotherapy (Table 3).
Gene Expression of BCL2 Exhibits the Expected Relationships in an Expanded Population
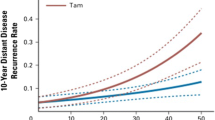
In the multivariate model, increased BCL2 expression was associated with a negative clinical outcome, that suggest a positive relationship with prognosis, which appears contrary to previous reports [30–32]. To confirm absence of bias in the measurement of BCL2, qPCR results for BCL2 gene expression in each carcinoma specimen were compared to ER status, grade, stage, and RFS in the entire qPCR dataset (Fig. 3). As described earlier, this patient population includes ER-negative and later-stage breast cancers. The median level of BCL2 gene expression was greater in ER-positive cancers compared to ER-negative cancers, confirming previous reports [32]. Furthermore, the median level of BCL2 expression was lower in higher-grade and later-stage cancers. When the median of BCL2 expression levels of 279 specimens was applied as a cutoff, increased BCL2 mRNA levels were associated with longer RFS of breast cancer patients, in agreement with previous reports [30–32]. Independently, BCL2 mRNA levels exhibited the expected association with clinical parameters, including RFS. The negative association between BCL2 gene expression breast cancers and RFS/OS was observed only in the multivariate model developed using ER+/PR+, early-stage breast cancers treated with adjuvant tamoxifen. In our opinion, these results suggest the possibility of a novel relationship between BCL2 and RFS in this ER+/PR+, tamoxifen-treated population of breast cancer patients, which may be explained in part by its biological interaction with ERBB4.
Relationships between BCL2 gene expression and clinicopathologic parameters of 279 primary breast carcinoma patients. Median BCL2 gene expression was compared between ER-negative and ER-positive breast cancers (a) by the Mann–Whitney test (P < 0.001). Similar comparisons were performed for tumor grade (b) and stage (c) using the Kruskal–Wallis test (P < 0.001 and P = 0.001, respectively). BCL2 expression was correlated with RFS by Kaplan–Meier regression (d). Logrank test revealed a hazard ratio (95% CI) of 1.91 (1.17–3.09) for decreased BCL2 expression (P = 0.001)
Discussion
Following NCI conferences relating the presence of ER and PR in breast cancer to clinical outcome [33], the NSABP conducted clinical trials administering adjuvant tamoxifen [23, 24]. After gathering biochemical and clinical data, NSABP investigations demonstrated the relationship between clinical outcome of tamoxifen-treated breast cancer and levels of either ER or PR in primary breast cancer biopsies. Decades of studies (e.g., [1]) established that while ER is a weak predictive index of prognosis, its clinical utility is significant in predicting a breast cancer patient’s response to tamoxifen administration.
With widespread use of tamoxifen for ER-positive breast cancers, it was recognized that a number of these patients have breast cancers refractory to this therapy (e.g., [1, 14]). In addition, PR expression is of clinical relevance (e.g., [12]), in that expression of only one of the sex-hormone receptors is associated with increased risk of failing tamoxifen therapy. The goal of our investigation was to use gene expression profiling to identify a sub-population of unresponsive breast cancer patients among those considered to be the most likely to respond to tamoxifen, i.e., women with early-stage, ER+/PR+ breast cancers. The results of this study, while significant, warrant further investigation in a larger retrospective study.
Using 12 candidate genes, we developed a five-gene model for predicting recurrence in patients treated with adjuvant tamoxifen for stages 1–2 breast cancers that are ER+/PR+. In general, these patients are considered at low-risk of recurrence [1, 10, 14]. It is proposed that collective expression of this gene set composed of PgR, BCL2, ERBB4 JM-a, RERG, and CD34 provides a molecular tool for identifying patients at a high risk for breast cancer recurrence. This gene expression model stratified the training population according to RFS and OS with similar significance observed using an independent patient population for validation. Furthermore, focusing on a population of early-stage breast cancers positive for both ER and PR is an important distinction due to the prognostic significance of PR expression in tamoxifen-treated patients. Patients with these types of breast cancers are most likely to respond to tamoxifen therapy, and the gene expression model we describe identifies refractory breast cancers within this group that would have otherwise been misclassified according to standard clinical parameters.
Other investigators have reported gene expression profiles in breast cancer from patients treated with tamoxifen (e.g., [16, 17]). A study by Kok et al. compared a number of gene expression tests available using “an independent cohort of (neo)adjuvant systemic treatment naïve breast cancer patients treated with first-line tamoxifen for metastatic disease” [34]. Although they reported that there was a low concordance of results among the three gene expression tests examined, in that only 45–61% of patients were classified in the same category, inclusion of gene signatures was recommended with IHC assays of ER. Unlike our investigation, the report from Kok et al. describes time to progression of tamoxifen-naïve patients who have already had a metastasis, whereas we examined a population of patients with early-stage lesions given tamoxifen for their primary cancer. In agreement, their investigation and ours incorporated PR substantiating the importance of considering both estrogen and progestin receptor levels when treating breast cancer patients with tamoxifen in either the adjuvant setting or after appearance of distant metastases.
Our multivariate analyses revealed expression of PgR, ERBB4 JM-a, RERG, and CD34 in breast cancer was positively associated with RFS. In contrast, increased BCL2 expression exhibited a negative association with RFS in this model. BCL2 is a member of a family of pro- or anti-apoptotic proteins that influence release of cytochrome c from mitochondria. Despite its anti-apoptotic function, BCL2 expression has been shown to have a positive association with breast cancer prognosis (e.g., [30, 32]), low tumor grade, and ER positivity (e.g., [32]). Callagy et al. [30] performed a meta-analysis (5,892 total cases) and concluded absence of BCL2 protein expression in primary breast cancers was associated with decreased RFS and OS. BCL2 gene expression was also reported to be an independent prognostic marker for breast cancer [31] and is included in the Oncotype Dx platform (e.g., [16, 17]). Our results suggest we have identified a novel relationship between BCL2 gene expression and clinical outcome in the context of this multivariate model. In experimental models, tamoxifen appears to induce cytochrome c release from mitochondria [35, 36] as well as downregulate expression of BCL2 [37, 38]. When BCL2 expression is already decreased, these effects may be enhanced by tamoxifen, which may explain the positive clinical outcome (i.e., increased RFS and OS) associated with decreased BCL2 expression in our multivariate model for tamoxifen-treated patients. The clinical significance of BCL2 expression in this model may also be related to the expression of the other genes, including ERBB4.
ERBB4 is one of the four receptor proteins in the ErbB receptor tyrosine kinase family. Characteristics of ERBB4 are unique in that activity of one of its splice variants, JM-a, encodes a 23 amino acid sequence that is susceptible to proteolytic cleavage, allowing release of an intracellular cytoplasmic domain (4ICD) [39]. In cultured breast cancer cells, 4ICD appears to translocate into mitochondria and promote apoptosis as a BH-3 only protein inhibiting anti-apoptotic function of BCL2 [40]. Furthermore, a ER–ERBB4-4ICD complex has been reported to influence transcription of certain genes via an estrogen response element (ERE) half-site, allowing for an autocrine mechanism increasing ERBB4 expression, as well as that of PgR [41]. Tamoxifen treatment of breast cancer cells apparently disrupts the ER–4ICD transcriptional coactivator complex and promotes activation of intrinsic apoptotic signaling by 4ICD, thereby enhancing BCL2 inhibition by ERBB4 [28]. Furthermore, 4ICD expression has been reported as a possible protein biomarker predicting breast cancer response to tamoxifen treatment [28]. The inhibitory effect of ERBB4 on BCL2 anti-apoptotic activity (enhanced by tamoxifen treatment) may explain the opposing relationships between their gene expression and RFS/OS in this tamoxifen-treated population.
Ras-like estrogen-regulated growth inhibitor (RERG) is a protein in the ras family with intrinsic GTP hydrolase activity but lacks a prenylation signal causing cytoplasmic, rather than membrane localization [42]. Finlin et al. reported RERG gene expression was stimulated by estradiol and inhibited by tamoxifen, indicating likely regulation by ER alpha [42]. Using microarray analyses, they also suggested RERG gene expression was associated with a breast cancer subtype exhibiting low proliferation and ER positivity. Similarly, we previously described RERG gene expression in an ER-positive molecular subtype associated with favorable clinical outcome [19]. The positive association between RERG expression and RFS in our multivariate model is consistent with results obtained in these previous studies.
CD34 is a cell surface glycoprotein found on certain hematopoietic stem cells with the capability of reconstituting all hematopoietic lineages [43]. The CD34 gene is estrogen-responsive with a high-affinity ERE sequence [44]. In breast cancer, CD34 expression is considered an immunohistochemical marker of angiogenesis, apparently correlating with decreased prognosis in patients not receiving systemic therapy [45]. In our multivariate analyses, CD34 gene expression had a positive association with the clinical outcome of patients treated with adjuvant tamoxifen. This clinical correlation may be explained by the biological function of CD34 (i.e., increased angiogenesis) which could enhance drug delivery to the lesion.
Using qPCR measurements of gene expression, a five-gene model was developed for assessment of prognosis in ER+/PR+, early-stage breast cancers treated with adjuvant tamoxifen. Collectively, these genes, each of which contains a known or candidate ERE sequence in the 5′ flanking region, appear to play roles in estrogen signaling pathways critical for breast cancer progression and response to endocrine therapy. Furthermore, there are direct biological interactions of the protein products of ERBB4 JM-a with those of BCL2 and PgR in breast cancer cells affecting apoptosis, cell growth, and mechanisms of tamoxifen response, which may be the molecular basis for the clinical relevance of expression of these genes identified in the multivariate model. Overall, this unique five-gene subset classified tamoxifen-treated patients with early-stage, ER+/PR + lesions who are at high risk for breast cancer recurrence although their clinicopathologic features indicated a favorable outcome.
References
Cleator SJ, Ahamed E, Coombes RC, Palmieri C (2009) A 2009 update on the treatment of patients with hormone receptor-positive breast cancer. Clin Breast Cancer 9(Suppl 1):S6–S17
Fleisher M, Dnistrian AM, Sturgeon CM, Wittliff JL (2002) Practice guidelines and recommendations for use of tumor markers in the clinic. In: Diamandis DP, Fritsche HA, Lilja H, Chan DW, Schwartz MK (eds) Tumor markers: physiology, pathobiology, technology and clinical applications. AACC, Washington, DC, pp 33–63
Sturgeon CM, Duffy MJ, Stenman UH et al (2008) National Academy of Clinical Biochemistry laboratory medicine practice guidelines for use of tumor markers in testicular, prostate, colorectal, breast, and ovarian cancers. Clin Chem 54:e11–e79
Wittliff JL, Pasic R, Bland KI (1998) Steroid and peptide hormone receptors: methods, quality control, and clinical use. In: Bland KI, Copeland EM (eds) The breast: comprehensive management of benign and malignant diseases. Saunders, Philadelphia, pp 458–498
Harris L, Fritsche H, Mennel R et al (2007) American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol 25:5287–5312
Bezwoda WR, Esser JD, Dansey R, Kessel I, Lange M (1991) The value of estrogen and progesterone receptor determinations in advanced breast cancer. Estrogen receptor level but not progesterone receptor level correlates with response to tamoxifen. Cancer 68:867–872
Dowsett M, Houghton J, Iden C et al (2006) Benefit from adjuvant tamoxifen therapy in primary breast cancer patients according oestrogen receptor, progesterone receptor, EGF receptor and HER2 status. Ann Oncol 17:818–826
Payne SJ, Bowen RL, Jones JL, Wells CA (2008) Predictive markers in breast cancer—the present. Histopathology 52:82–90
Wittliff JL (1984) Steroid-hormone receptors in breast cancer. Cancer 53:630–643
Grann VR, Troxel AB, Zojwalla NJ, Jacobson JS, Hershman D, Neugut AI (2005) Hormone receptor status and survival in a population-based cohort of patients with breast carcinoma. Cancer 103:2241–2251
Ravdin PM, Green S, Dorr TM et al (1992) Prognostic significance of progesterone receptor levels in estrogen receptor-positive patients with metastatic breast cancer treated with tamoxifen: results of a prospective Southwest Oncology Group study. J Clin Oncol 10:1284–1291
Bardou VJ, Arpino G, Elledge RM, Osborne CK, Clark GM (2003) Progesterone receptor status significantly improves outcome prediction over estrogen receptor status alone for adjuvant endocrine therapy in two large breast cancer databases. J Clin Oncol 21:1973–1979
Rakha EA, El-Sayed ME, Green AR et al (2007) Biologic and clinical characteristics of breast cancer with single hormone receptor positive phenotype. J Clin Oncol 25:4772–4778
Bryant J, Fisher B, Dignam J (2001) Duration of adjuvant tamoxifen therapy. J Natl Cancer Inst Monogr 30:56–61
Clark GM, McGuire WL (1983) Progesterone receptors and human breast cancer. Breast Cancer Res Treat 3:157–163
Paik S, Shak S, Tang G et al (2004) A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 351:2817–2826
Sparano JA, Paik S (2008) Development of the 21-gene assay and its application in clinical practice and clinical trials. J Clin Oncol 26:721–728
Vendrell JA, Robertson KE, Ravel P et al (2008) A candidate molecular signature associated with tamoxifen failure in primary breast cancer. Breast Cancer Res 10(5):R88
Ma XJ, Wang W, Salunga R et al (2003) Gene expression associated with clinical outcome in breast cancer via laser capture microdissection. Breast Cancer Res Treat 82:A29
Wittliff JL, Erlander MG (2002) Laser capture microdissection and its applications in genomics and proteomics. Methods Enzymol 356:12–25
Jin VX, Sun H, Pohar TT et al (2005) ERTargetDB: an integral information resource of transcription regulation of estrogen receptor target genes. J Mol Endocrinol 35(2):225–230
Fitzgibbons PL, Page DL, Weaver D et al (2000) Prognostic factors in breast cancer. College of American Pathologists Consensus Statement 1999. Arch Pathol Lab Med 124:966–978
Fisher B, Redmond C, Brown A et al (1981) Treatment of primary breast cancer with chemotherapy and tamoxifen. N Engl J Med 305:1–6
Fisher B, Redmond C, Brown A et al (1983) Influence of tumor estrogen and progesterone receptor levels on the response to tamoxifen and chemotherapy in primary breast cancer. J Clin Oncol 1:227–241
McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM (2005) Reporting recommendations for tumor marker prognostic studies. J Clin Oncol 23:9067–9072
Schroeder A, Mueller O, Stocker S et al (2006) The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol 7:3
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45
Naresh A, Thor AD, Edgerton SM, Torkko KC, Kumar R, Jones FE (2008) The HER4/4ICD estrogen receptor coactivator and BH3-only protein is an effector of tamoxifen-induced apoptosis. Cancer Res 68:6387–6395
Dupont WD (2002) Statistical modeling for biomedical researchers: a simple introduction to the analysis of complex data. Cambridge University Press, Cambridge
Callagy GM, Webber MJ, Pharoah PD, Caldas C (2008) Meta-analysis confirms BCL2 is an independent prognostic marker in breast cancer. BMC Cancer 8:153
Thomadaki H, Talieri M, Scorilas A (2007) Prognostic value of the apoptosis related genes BCL2 and BCL2L12 in breast cancer. Cancer Lett 247:48–55
Callagy GM, Pharoah PD, Pinder SE et al (2006) Bcl-2 is a prognostic marker in breast cancer independently of the Nottingham Prognostic Index. Clin Cancer Res 12:2468–2475
DeSombre ER, Carbone PP, Jensen EV et al (1979) Special report. Steroid receptors in breast cancer. N Engl J Med 301:1011–1012
Kok M, Linn SC, Van Laar RK et al (2009) Comparison of gene expression profiles predicting progression in breast cancer patients treated with tamoxifen. Breast Cancer Res Treat 113(2):275–283
Kallio A, Zheng A, Dahllund J, Heiskanen KM, Harkonen P (2005) Role of mitochondria in tamoxifen-induced rapid death of MCF-7 breast cancer cells. Apoptosis 10:1395–1410
Obrero M, Yu DV, Shapiro DJ (2002) Estrogen receptor-dependent and estrogen receptor-independent pathways for tamoxifen and 4-hydroxytamoxifen-induced programmed cell death. J Biol Chem 277:45695–45703
Diel P, Smolnikar K, Michna H (1999) The pure antiestrogen ICI 182780 is more effective in the induction of apoptosis and down regulation of BCL-2 than tamoxifen in MCF-7 cells. Breast Cancer Res Treat 58:87–97
Thiantanawat A, Long BJ, Brodie AM (2003) Signaling pathways of apoptosis activated by aromatase inhibitors and antiestrogens. Cancer Res 63:8037–8050
Rio C, Buxbaum JD, Peschon JJ, Corfas G (2000) Tumor necrosis factor-alpha-converting enzyme is required for cleavage of erbB4/HER4. J Biol Chem 275:10379–10387
Naresh A, Long W, Vidal GA et al (2006) The ERBB4/HER4 intracellular domain 4ICD is a BH3-only protein promoting apoptosis of breast cancer cells. Cancer Res 66:6412–6420
Zhu Y, Sullivan LL, Nair SS et al (2006) Coregulation of estrogen receptor by ERBB4/HER4 establishes a growth-promoting autocrine signal in breast tumor cells. Cancer Res 66:7991–7998
Finlin BS, Gau CL, Murphy GA et al (2001) RERG is a novel ras-related, estrogen-regulated and growth-inhibitory gene in breast cancer. J Biol Chem 276:42259–42267
Sutherland DR, Keating A (1992) The CD34 antigen: structure, biology, and potential clinical applications. J Hematother 1:115–129
Bourdeau V, Deschenes J, Metivier R et al (2004) Genome-wide identification of high-affinity estrogen response elements in human and mouse. Mol Endocrinol 18:1411–1427
Dhakal HP, Naume B, Synnestvedt M et al (2008) Vascularization in primary breast carcinomas: its prognostic significance and relationship with tumor cell dissemination. Clin Cancer Res 14:2341–2350
Acknowledgments
Authors acknowledge the technical assistance of Dr. Irina A. Smolenkova and advice of Traci L. Kruer and Dr. Sarah A. Andres, Department of Biochemistry, and Dr. Guy Brock, Department of Bioinformatics and Biostatisticis, for assistance with statistical analyses. This study was supported in part by grants from the Phi Beta Psi Sorority Charity Trust and a CTSP award from the Commonwealth of Kentucky. DAK was a recipient of a Graduate Fellowship from the Integrated Programs in Biomedical Sciences, University of Louisville, and the Spatola Graduate Fellowship from IMD3.
Conflicts of Interest
Financial disclosures: A patent has been filed (US patent application no. 13/123,158) in the name of the University of Louisville with James L. Wittliff and D. Alan Kerr II as co-inventors. This patent includes some of the data presented in this manuscript regarding gene expression patterns predicting recurrence in tamoxifen-treated patients. There are no consultancies, stock ownerships, or any other financial relationships involving the authors that would bias the work presented in this manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kerr, D.A., Wittliff, J.L. A Five-Gene Model Predicts Clinical Outcome in ER+/PR+, Early-Stage Breast Cancers Treated with Adjuvant Tamoxifen. HORM CANC 2, 261 (2011). https://doi.org/10.1007/s12672-011-0080-8
Published:
DOI: https://doi.org/10.1007/s12672-011-0080-8