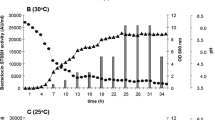
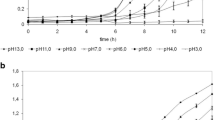
Abstract
Total DNA extracted from Lb. plantarum ST8Sh was screened for the presence of more than 50 genes related to production of biogenic amines (histidine decarboxylase, tyrosine decarboxylase, and ornithine decarboxylase), virulence factors (sex pheromones, gelatinase, cytolysin, hyaluronidase, aggregation substance, enterococcal surface protein, endocarditis antigen, adhesion of collagen, integration factors), and antibiotic resistance (vancomycin, tetracycline, erythromycin, gentamicin, chloramphenicol, bacitracin). Lb. plantarum ST8Sh showed a low presence of virulence genes. Only 13 genes were detected (related to sex pheromones, aggregation substance, adhesion of collagen, tetracycline, gentamicin, chloramphenicol, erythromycin, but not to vancomycin, and bacitracin) and may be considered as indication of safety for application in fermented food products. In addition, interaction between Lb. plantarum ST8Sh and drugs from different groups were determined in order to establish possible application of the strain in combination with commercial drugs. Cytotoxicity of the semi-purified bacteriocins produced by Lb. plantarum ST8Sh was depended on applied concentration—highly cytotoxic when applied at 25 μg/mL and no cytotoxicity at 5 μg/mL.
Similar content being viewed by others
References
Martinez RCR, Vieira ADS, Santos KMO, Franco BDGM, Todorov SD (2012) Characterization and evaluation of Lactobacillus plantarum probiotic potential. In: AIP C, Mena AL (eds) Lactobacillus: classification, uses and health implications, Bacteriology Research Developments / Microbiology Research Advances series. Nova Publishers, New York, USA, pp 36–63
Doern CD, Nguyen ST, Afolabi F, Burnham C-AD (2014) Probiotic-associated aspiration pneumonia due to Lactobacillus rhamnosus. J Clin Microbiol 52:3124–3126
Goldstein EJC, Tyrrell KL, Citron DM (2015) Lactobacillus species: taxonomic complexity and controversial susceptibilities. Clin Infect Dis 60:S98–S107
Schwendicke F, Dörfer C, Kneist S, Meyer-Lueckel H, Paris S (2014) Cariogenic effects of probiotic Lactobacillus rhamnosus GG in a dental biofilm model. Caries Res 48:186–192
Dicks LMT, Todorov SD, Franco BDGM (2011) Current status of antibiotic resistance in lactic acid bacteria. In: Bonilla AR, Muniz KP (eds) Antibiotic resistance: causes and risk factors, mechanisms and alternatives. Pharmacology—research, safety testing and regulation. Nova Publisher, New York, USA, pp 379–425
Todorov SD, Botes M, Danova ST, Dicks LMT (2007) Probiotic properties of Lactococcus lactis subsp. lactis HV219, isolated from human vaginal secretions. J Appl Microbiol 103:629–639
Todorov SD, Botes M, Guigas C, Schillinger U, Wiid I, Wachsman MB, Holzapfel WH, Dicks LMT (2008) Boza, a natural source of probiotic lactic acid bacteria. J Appl Microbiol 104:465–477
De Carvalho KG, Kruger MF, Furtado DN, Todorov SD, Franco BDGM (2009) Evaluation of the role of environmental factors in the human gastrointestinal tract on the behaviour of probiotic cultures Lactobacillus casei Shirota and Lactobacillus casei LC01 by the use of a semi-dynamic in vitro model. Ann Microbiol 59:439–445
Bogsan CS, Nero LA, Todorov SD (2015) From traditional knowledge to an innovative approach for bio-preservation in food by using lactic acid bacteria.Chapter 01. In: Liong M-T (ed) Beneficial microorganisms in Food & Nutraceuticals. Springer, New York, NY, USA, pp 1–36
Jozala AF, De Andrade MS, De Arauz LJ, Pessoa A Jr, Penna TCV (2007) Nisin production utilizing skimmed milk aiming to reduce process cost. Appl Biochem Biotechnol 137-140:515–528
Deegan LH, Cotter PD, Hill C, Ross P (2006) Bacteriocins: biological tools for bio-preservation and shelf-life extension. Int Dairy J 16:1058–1071
Motta AS, Cannavan FS, Tsai S-M, Brandelli A (2007) Characterization of a broad range antibacterial substances from a new Bacillus species isolated from Amazon basin. Arch Microbiol 188:367–375
Vaucher AR, Gewehr CVC, Correa FAP, Sant'Anna V, Ferreira J, Brandelli A (2011) Evaluation of the immunogenicity and in vivo toxicity of the antimicrobial peptide P34. Int J Pharm 421:94–98
Klaenhammer TR (1988) Bacteriocins of lactic acid bacteria. Biochimie 70:337–349
Todorov SD, Holzapfel W, Nero LA (2016) Characterization of a novel bacteriocin produced by Lactobacillus plantarum ST8SH and some aspects of its mode of action. Ann Microbiol 66:949–962
Todorov SD, Holzapfel WH, Nero LA (2016) In Vitro evaluation of beneficial properties of bacteriocinogenic Lactobacillus plantarum ST8Sh. Probiotics & Antimicrobial Proteins. doi:10.1007/s12602-016-9245-7. (in press)
Moraes PM, Perin LM, Todorov SD, Silva A Jr, Franco BDGM, Nero LA (2012) Bacteriocinogenic and virulence potential of Enterococcus isolates obtained from raw milk and cheese. J Appl Microbiol 113:318–328
Fortina MG, Ricci G, Borgo F, Manachini PL, Arends K, Schiwon K, Abajy MY, Grohmann E (2008) A survey on biotechnological potential and safety of the novel Enterococcus species of dairy origin, E. italicus. Int J Food Microbiol 123:204–211
Barbosa J, Gibbs PA, Teixeira P (2010) Virulence factors among enterococci isolated from traditional fermented meat products produced in the north of Portugal. Food Control 21:651–656
Eaton TJ, Gasson MJ (2001) Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Applied and environmental Microbiology 67:1628–1635
Favaro L, Basaglia M, Casella S, Hue I, Dousset X, Franco BDGM, Todorov SD (2004) Bacteriocinogenic potential and safety evaluation of non-starter enterococcus faecium strains isolated from home made white brine cheese. Food Microbiol 38:228–239
Favaro L, Corich V, Giacomini A, Basaglia M, Casella S (2013) Grape marcs as unexplored source of new yeasts for future biotechnological applications. World J Microbiol Biotechnol 29:1551–1562
Smith PB, Hancock GA, Rhoden DL (1969) Improved medium for detecting deoxyribonuclease-producing bacteria. Appl Microbiol 18:991–993
Carneiro BM, Braga ACS, Batista MN, Rahal P, Favaro L, Penna ALB, Todorov SD (2014) Lactobacillus plantarum ST202Ch and Lactobacillus plantarum ST216Ch—what are the limitations for application? Journal of Nutritional Health and Food Engineering 1(2):00010
Vaucher RA, Teixeira ML, Brandelli A (2010) Investigation of the cytotoxicity of antimicrobial peptide P40 on eukaryotic cells. Curr Microbiol 60:1–5
Gomes BC, Esteves CT, Palazzo ICV, Darini ALC, Felis GE, Sechi LA, Franco BDGM, de Martinis ECP (2008) Prevalence and characterization of Enterococcus spp. isolated from Brazilian foods. Food Microbiol 25:668–675
Teuber M (1999) Spread of antibiotic resistance with food-borne pathogens. Cell Molecular Life Science 56:755–763
Todorov SD, Franco BDGM, Wiid IJ (2013) In vitro study of beneficial properties and safety of lactic acid bacteria isolated from Portuguese fermented meat products. Benefic Microbes 5:351–366
De Sousa CP (2003) Pathogenicity mechanisms of prokaryotic cells: an evolutionary view. Braz J Infect Dis 7:23–31
Franz CM, Muscholl-Silberhorn AB, Yousif NMK, Vancanneyt M, Swings J, Holzapfel WH (2001) Incidence of virulence factors and antibiotic resistance among enterococci isolated from food. Appl Environ Microbiol 67:4385–4389
Todorov S, Wachsman M, Ts I-I, Ivanova I (2014) Lactobacillus plantarum ST16Pa—are we ready to use it as bio-preservative culture? Bulgarian Journal of Agricultural Science 20:55–58
Holzapfel WH, Wood BJB (2014) Introduction to the LAB. Chapter 1. In: Holzapfel WH, Wood BJB (eds) Lactic acid bacteria—biodiversity and taxonomy. John Wiley & Sons Ltd., Chichester, West Sussex, UK, pp 1–12
Franz CMAP, Stiles ME, Schleifer K-H, Holzapfel WH (2003) Enterococci in foods—a conundrum for food safety. Int J Food Microbiol 88:105–122
Hendrickx APA, Willems RJL, Bonten MJM, van Schaik W (2009) LPxTG surface proteins of enterococci. Trends Microbiol 17:423–430
Franz CMAP, Holzapfel WH (2004) The genus Enterococcus: biotechnological and safety issues. In: Salminen S, von Wright A, Ouwehand A (eds) The lactic acid bacteria: microbiology and functional aspects, 3rd edn. New York, Marcel Dekker, pp 199–247
Cook LC, Federle MJ (2014) Peptide pheromone signaling in Streptococcus and Enterococcus. FEMS Microbiol Rev 38:473–492
Botes M, Van Reenen CA, Dicks LMT (2008) Evaluation of Enterococcus mundtii ST4SA and Lactobacillus plantarum 423 as probiotics by using a gastro-intestinal model with infant milk formulations as substrate. Int J Food Microbiol 128:362–370
Todorov SD, Dicks LMT (2008) Evaluation of lactic acid bacteria from kefir, molasses and olive brine as possible probiotics based on physiological properties. Ann Microbiol 58:661–670
Ammor MS, Flórez AB, Mayo B (2007) Antibiotic resistance in non-enterococcal lactic acid bacteria and bifidobacteria. Food Microbiol 24:559–570
Levy SB, Marshall B (2004) Antibacterial resistance worldwide: causes, challenges and responses. Nat Med 10:S122–S129
Salyers AA, Gupta A, Wang Y (2004) Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol 12:412–416
Danielsen M, Wind A (2003) Susceptibility of Lactobacillus spp. to antimicrobial agents. Int J Food Microbiol 82:1–11
Elisha BG, Courvalin P (1995) Analysis of genes encoding d-alanine: d-alanine ligase-related enzymes in Leuconostoc mesenteroides and Lactobacillus spp. Gene 152:79–83
Salminen MK, Rautelin H, Tynkkynen S, Poussa T, Saxelin M, Valtonen V, Jarvinen A (2006) Lactobacillus bacteremia, species identification, and antimicrobial susceptibility of 85 blood isolates. Clin Infect Dis 42:35–44
Charteris WP, Kelly PM, Morelli L, Collins JK (1998) Development and application of an in vitro methodology to determine the transit tolerance of potentially probiotic Lactobacillus and Bifidobacterium species in theupper human gastrointestinal tract. Jornal of Applied Microbiolog 84:759–768
Coppola R, Succi M, Tremonte P, Reale A, Salzano G, Sorrentino Z (2005) Antibiotic susceptibility of Lactobacillus rhamnosus strains isolated from Parmegiano Reggiona cheese. Lait 85:193–204
Zhou JS, Pillidge CJ, Gopal PK, Gill HS (2005) Antibiotic susceptibility profiles of new probiotic Lactobacillus and Bifidobacterium strains. Int J Food Microbiol 98:211–217
Herreros MA, Sandoval H, González L, Castro JM, Fresno JM, Tornadijo ME (2005) Antimicrobial activity and antibiotic resistance of lactic acid bacteria isolated from armada cheese (a Spanish goats’ milk cheese). Food Microbiol 22:455–459
Herrero M, Mayo B, Ganzales B, Suarez JE (1996) Evaluation of technologically important traits in lactic acid bacteria isolated from spontaneous fermentations. J Appl Bacteriol 81:565–570
Elkins CA, Mullis LB (2004) Bile-mediated aminoglycoside sensibility in Lactobacillus species likely results from increased membrane permeability attributable to cholic acid. Appl Environ Microbiol 70:7200–7209
Rojo-Bezares B, Sáenz Y, Poeta P, Zarazaga M, Ruiz-Larrea F, Torres C (2006) Assessment of antibiotic susceptibility within lactic acid bacteria strains isolated from wine. Int J Food Microbiol 111:234–240
Ahn C, Collins-Thompson D, Duncan C, Stiles ME (1992) Mobilization and location of the genetic determinant of chloramphenicol resistance from Lactobacillus plantarum caTC2R. Plasmid 27:169–176
Huys G, D’Haene K, Swings J (2006) Genetic basis of tetracycline and minocycline resistance in potentially probiotic Lactobacillus plantarumstrain CCUG 43738. Antimicrob Agents Chemother 50:1550–1551
Salyers AA, Shoemaker NB (1996) Resistance gene transfer in anaerobes: new insights, new problems. Clin Infect Dis 23:S36–S43
dos Santos KMO, Vieira ADS, Buriti FCA, do Nascimento JCF, de Melo MES, Bruno LM, Borges MF, Rocha CRC, Lopes ACS, Franco BDGM, Todorov SD (2015) Artisanal Coalho cheeses as source of beneficil Lactobacillus plantarum and Lactobacillus rhamnosus strains. Dairy Science & Technology 95:209–230
Acknowledgements
This work has been supported by research and personal grants from FAPEMIG (Belo Horizonte, MG, Brazil), CAPES (Brasilia, DF, Brazil), and CNPq (Brasilia, DF, Brazil). To Braga A.C.S. and Batista M.N. from UNESP — Universidade Estadual Paulista, Instituto de Biociências, Letras e Ciências Exatas. Departamento de Engenharia e Tecnologia de Alimentos, São José do Rio Preto, SP, Brazil for technical assistance related to cytotoxicity of bacteriocin work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Todorov, S.D., Perin, L.M., Carneiro, B.M. et al. Safety of Lactobacillus plantarum ST8Sh and Its Bacteriocin. Probiotics & Antimicro. Prot. 9, 334–344 (2017). https://doi.org/10.1007/s12602-017-9260-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12602-017-9260-3