Abstract
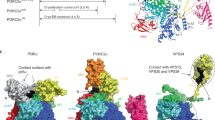
PI3Kα, a heterodimeric lipid kinase, catalyzes the conversion of phosphoinositide-4,5-bisphosphate (PIP2) to phosphoinositide-3,4,5-trisphosphate (PIP3), a lipid that recruits to the plasma membrane proteins that regulate signaling cascades that control key cellular processes such as cell proliferation, carbohydrate metabolism, cell motility, and apoptosis. PI3Kα is composed of two subunits, p110α and p85, that are activated by binding to phosphorylated receptor tyrosine kinases (RTKs) or their substrates. The gene coding for p110α, PIK3CA, has been found to be mutated in a large number of tumors; these mutations result in increased PI3Kα kinase activity. The structure of the complex of p110α with a fragment of p85 containing the nSH2 and the iSH2 domains has provided valuable information about the mechanisms underlying the physiological activation of PI3Kα and its pathological activation by oncogenic mutations. This review discusses information derived from x-ray diffraction and theoretical calculations regarding the structural and dynamic effects of mutations in four highly mutated regions of PI3K p110α, as well as the proposed mechanisms by which these mutations increase kinase activity. During the physiological activation of PI3Kα, the phosphorylated tyrosine of RTKs binds to the nSH2 domain of p85, dislodging an inhibitory interaction between the p85 nSH2 and a loop of the helical domain of p110α. Several of the oncogenic mutations in p110α activate the enzyme by weakening this autoinhibitory interaction. These effects involve structural changes as well as changes in the dynamics of the enzyme. One of the most common p110α mutations, H1047R, activates PI3Kα by a different mechanism: it increases the interaction of the enzyme with the membrane, maximizing the access of the PI3Kα to its substrate PIP2, a membrane lipid.




Similar content being viewed by others
References
Atilgan A, Durell S et al (2001) Anisotropy of fluctuation dynamics of proteins iwth an elastic network model. Biophys J 80(1):505–515
Bachman KE, Argani P et al (2004) The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther 3(8):772–775
Berndt A, Miller S et al (2010) The p110delta structure: mechanisms for selectivity and potency of new PI(3)K inhibitors. Nat Chem Biol 6(3):244
Broderick DK, Di C et al (2004) Mutations of PIK3CA in anaplastic oligodendrogliomas, high-grade astrocytomas, and medulloblastomas. Cancer Res 64(15):5048–5050
Burke JE, Perisic O et al (2012) Oncogenic mutations mimic and enhance dynamic events in the natural activation of phosphoinositide 3-kinase p110alpha (PIK3CA). Proc Natl Acad Sci USA 109(38):15259–15264In this paper the dynamics of PI3K are analyzed by hydrogen deuterium exchange mass spectrometry. It Identifies regions of enhanced dynamics in the heterodimer.
Burke JE, Vadas O et al (2011) Dynamics of the phosphoinositide 3-kinase p110delta interaction with p85alpha and membranes reveals aspects of regulation distinct from p110alpha. Structure 19(8):1127–1137
Campbell IG, Russell SE et al (2004) Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res 64(21):7678–7681
Carson JD, Van Aller G et al (2008) Effects of oncogenic p110alpha subunit mutations on the lipid kinase activity of phosphoinositide 3-kinase. Biochem J 409(2):519–524
Certal V, Halley F et al (2012) Discovery and optimization of new benzimidazole- and benzoxazole-pyrimidone selective PI3Kbeta inhibitors for the treatment of phosphatase and TENsin homologue (PTEN)-deficient cancers. J Med Chem 55(10):4788–4805
Cgarn CGARN (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455(7216):1061–1068
Dhand R, Hara K et al (1994) PI 3-kinase: structural and functional analysis of intersubunit interactions. EMBO J 13(3):511–521
Engelman JA, Luo J et al (2006) The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet 7(8):606–619
Eyal, E., A. Dutta, et al. (2011) Cooperative dynamics of proteins unraveled by network models. WIREs Computational Molecular Science 1 (May-June)
Forbes SA, Bindal N et al (2011) COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer.. Nucleic Acids Res 39(Database issue):D945–D950
Gabelli SB, Mandelker D et al (2010) Somatic mutations in PI3Kalpha: structural basis for enzyme activation and drug design. Biochim Biophys Acta 1804(3):533–540
Geering B, Cutillas PR et al (2007) Class IA phosphoinositide 3-kinases are obligate p85-p110 heterodimers. Proc Natl Acad Sci USA 104(19):7809–7814
Gur M, Zomot E et al (2013) Global motions exhibited by proteins in micro- to milliseconds simulations concur with anisotropic network model predictions. J Chem Phys 139(121912)
Gymnopoulos M, Elsliger MA et al (2007) Rare cancer-specific mutations in PIK3CA show gain of function. Proc Natl Acad Sci USA 104(13):5569–5574
Holt KH, Olson L et al (1994) Phosphatidylinositol 3-kinase activation is mediated by high-affinity interactions between distinct domains within the p110 and p85 subunits. Mol Cell Biol 14(1):42–49
Hon WC, Berndt A et al (2012) Regulation of lipid binding underlies the activation mechanism of class IA PI3-kinases. Oncogene 31(32):3655–3666
Huang CH, Mandelker D et al (2007) The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations. Science 318(5857):1744–1748 This paper describes the structure of the p110/nip85 heterodimer and identifies the high mutation regions in the structure.
Kang S, Bader AG et al (2004) Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci USA 102(3):802–807
Katso R, Okkenhaug K et al (2001) Cellular function of phosphoinositide 3-kinases: implications for development, homeostasis, and cancer. Annu Rev Cell Dev Biol 17:615–675
Lee JW, Soung YH et al (2005) PIK3CA gene is frequently mutated in breast carcinomas and hepatocellular carcinomas. Oncogene 24(8):1477–1480
Levine DA, Bogomolniy F et al (2005) Frequent mutation of the PIK3CA gene in ovarian and breast cancers. Clin Cancer Res 11(8):2875–2878
Mandelker D, Gabelli SB et al (2009) A frequent kinase domain mutation that changes the interaction between PI3Kalpha and the membrane. Proc Natl Acad Sci U S A 106(40):16996–17001
Miled N, Yan Y et al (2007) Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science 317(5835):239–242
Murray JM, Sweeney ZK et al (2012) Potent and highly selective benzimidazole inhibitors of PI3-kinase delta. J Med Chem 55(17):7686–7695
Nolte RT, Eck MJ et al (1996) “Crystal structure of the PI 3-kinase p85 amino-terminal SH2 domain and its phosphopeptide complexes”. Nat Struct Biol 3(4):364–374
Rudd ML, Price JC et al (2011) A unique spectrum of somatic PIK3CA (p110alpha) mutations within primary endometrial carcinomas. Clin Cancer Res 17(6):1331–1340
Samuels Y, Wang Z et al (2004) High frequency of mutations of the PIK3CA gene in human cancers. Science 304(5670):554
Shepherd PR, Siddle K et al (1997) Is stimulation of class-1 phosphatidylinositol 3-kinase activity by insulin sufficient to activate pathways involved in glucose metabolism. Biochem Soc Trans 25(3):978–981
Vanhaesebroeck B, Alessi DR (2000) The PI3K-PDK1 connection: more than just a road to PKB. Biochem J 346(Pt 3):561–576
Vanhaesebroeck B, Waterfield MD (1999) Signaling by distinct classes of phosphoinositide 3-kinases. Exp Cell Res 253(1):239–254
Vogt PK, Kang S et al (2007) Cancer-specific mutations in phosphatidylinositol 3-kinase. Trends Biochem Sci 32(7):342–349
Walker EH, Perisic O et al (1999) Structural insights into phosphoinositide 3-kinase catalysis and signalling. Nature 402(6759):313–320
Wu H, Yan Y et al (2007) Regulation of class IA PI3Ks. Biochem Soc Trans 35(Pt 2):242–244
Zhao L, Vogt PK (2008a) Class I PI3K in oncogenic cellular transformation. Oncogene 27(41):5486–5496
Zhao L, Vogt PK (2008b) Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci USA 105(7):2652–2657
Acknowledgments
This work was supported by the Virginia and D.K. Ludwig Fund for Cancer Research and by NIH grant CA43460. SBG is a Stewart Trust fellow.
Conflict of interest
Authors Sandra B. Gabelli, Ignacia Echeverria, Megan Alexander, Krisna C. Duong-Ly, Daniele Chaves-Moreira, Evan T. Brower, B. Vogelstein, and L. Mario Amzel declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by the any of the authors.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Special Issue Advances in Biophysics in Latin America
Rights and permissions
About this article
Cite this article
Gabelli, S.B., Echeverria, I., Alexander, M. et al. Activation of PI3Kα by physiological effectors and by oncogenic mutations: structural and dynamic effects. Biophys Rev 6, 89–95 (2014). https://doi.org/10.1007/s12551-013-0131-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12551-013-0131-1