Abstract
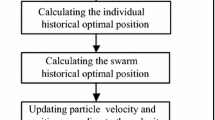
The primary structure of proteins consists of a linear chain of amino acids that can vary in length. Proteins fold, under the influence of several chemical and physical factors, into their 3D structures, which determine their biological functions and properties. Misfolding occurs when the protein folds into a 3D structure that does not represent its native structure, which can lead to diseases. Due to the importance of this problem and since laboratory techniques are not always feasible, computational methods for characterizing protein structures have been proposed. In this paper, we present a particle swarm optimization (PSO) based algorithm for predicting protein structures in the 3D hydrophobic polar model. Starting from a small set of candidate solutions, our algorithm efficiently explores the search space and returns 3D protein structures with minimal energy. To test our algorithm, we used two sets of benchmark sequences of different lengths and compared our results to published results. Our algorithm performs better than previous algorithms by finding lower energy structures or by performing fewer numbers of energy evaluations.
Similar content being viewed by others
References
Anfinsen, C.B. 1973. Principles that govern the folding of proteins. Science 181, 223–230.
Bastolla, U., Fravenkron, H., Gestner, E., Grassberger, P., Nadler, W. 1998. Testing a new Monte Carlo algorithm for the protein folding problem. Proteins 32, 52–66.
Bui, T.N., Sundarraj, G. 2005. An efficient genetic algorithm for predicting protein tertiary structures in the 2D HP model. In: Proceedings of the 2005 Conference on Genetic and Evolutionary Computation, USA, 385–392.
Chen, M., Huang, W. 2005. A branch and bound algorithm for the protein folding problem in the HP lattice model. Genomics Proteomics Bioinf 3, 225–230.
Cui, Y., Chen, R.S., Wong, W.H. 1998. Protein folding simulation with genetic algorithm and supersecondary structure constraints. Proteins 31, 247–257.
Custódio, F., Barbosa, H., Dardenne, L. 2004. Investigation of the three-dimensional lattice HP protein folding model using a genetic algorithm. Genet Mol Biol 27, 611–615.
Das, S., Abraham, A., Konar, A. 2008. Swarm intelligence algorithms in bioinformatics. In: Studies in Computational Intelligence. Springer, Berlin, 113–147.
Das, R., Baker, D. 2008. Macromolecular modeling with Rosetta. Annu Rev Biochem 77, 363–382.
Datta, A., Talukdar, V., Konar, A., Jain, L.C. 2008. Neuro-swarm hybridization for protein tertiary structure prediction. Int J Hybrid Intell Syst 5, 153–159.
Floudas, C. 2007. Computational methods in protein structure prediction. Biotechnol Bioeng 97, 207–213.
Hart, W.E., Newman, A. 2006. Protein structure prediction with lattice models. In: Aluru, S. (Ed.) Handbook of Molecular Biology, CRC Press, New York, 1–24.
Hsu, H.P., Mehra, V., Nadler, W., Grassberger, P. 2003. Growth algorithm for lattice heteropolymers at low temperatures. J Chem Phys 118, 444–451.
Johnson, C., Katikireddy, A. 2006. A genetic algorithm with backtracking for protein structure prediction. In: Proceedings of the th Annual Conference on Genetic and Evolutionary Computation, USA, 299–300.
Jones, D.T. 1998. THREADER: Protein sequence threading by double dynamic programming. In: Salzberg, S., Searl, D., Kasif, S. (Eds.) Computational Methods in Molecular Biology, Elsevier Science, Amsterdam, 285–312.
Kennedy, J., Eberhart, R.C. 1995. Particle swarm optimization. In: Proceedings of the 1995 IEEE International Conference of Neural Networks, Australia, 1942–1948.
Klepeis, J.L., Floudas, C.A. 2003. Ab initio tertiary structure prediction of proteins. J Global Optim 25, 113–140.
Kopp, J., Schwede, T. 2004. Automated protein structure homology modeling: A progress report. Pharm J 5, 405–416.
Lathrop, R.H., Rogers, R.G. Jr., Bienkowska, J., Bryant, B.K.M., Buturovic, L.J., Gaitatzes, C., Nambudripad, R., White, J.V., Smith, T.F. 1998. Analysis and algorithms for protein sequence-structure alignment. In: Salzberg, S.L., Searls, D.B., Kasif, S. (Eds.) Computational Methods in Molecular Biology, Elsevier Science, Amsterdam, 227–283.
Liang, F., Wong, W.H. 2001. Evolutionary Monte Carlo for protein folding simulations. J Chem Phys 115, 3374–3380.
Mansour, N., Kehyayan, C., Khachfe, H. 2009. Scatter search algorithm for protein structure prediction. Int J Bioinf Res Appl 5, 501–515.
Pandit, S.B., Zhang, Y., Skolnick, J. 2006. Tasser-lite: An automated tool for protein comparative modeling. Biophys J 91, 4180–4190.
Patton, A.L., Punch, W.F., Goodman, E.D. 1995. A standard GA approach to native protein conformation prediction. In: Proceedings of the Sixth International Conference on Genetic Algorithms, USA, 574–581.
Prusiner, S.B. 1998. Prions. Proc Natl Acad Sci USA 95, 13363–13383.
Roy, A., Kucukural, A., Zhang, Y. 2010. I-TASSER: A unified platform for automated protein structure and function prediction. Nat Protoc 5, 725–738.
Rylance, G. 2004. Applications of genetic algorithms in protein folding studies. First year report, School of Chemistry, University of Birmingham, England.
Schulze-Kremer, S. 2000. Genetic algorithms and protein folding. Methods Mol Biol 143, 175–222.
Shmygelska, A., Hoos, H.H. 2005. An Ant Colony optimization algorithm for the 2D and 3D hydrophobic polar protein folding problem. BMC Bioinf 6, 30.
Sikder, A.R., Zomaya, A.Y. 2005. An Overview of protein-folding techniques: issues and perspectives. Int J Bioinf Res Appl 1, 121–143.
Toma, L., Toma, S. 1996. Contact interactions method: A new algorithm for protein folding simulations. Protein Sci 5, 147–153.
Unger, R., Moult, J. 1993a. Finding the lowest free energy conformation of a protein is an NP-Hard problem: Proof and implications. Bull Math Biol 55, 1183–1198.
Unger, R., Moult, J. 1993b. Genetic algorithms for protein folding simulations. J Mol Biol 231, 75–81.
Wilke, D.N. 2005. Analysis of the Particle Swarm Optimization Algorithm. Master dissertation, University of Pretoria.
Yue, K., Dill, K.A. 1995. Forces of tertiary structural organization in globular proteins. Proc Natl Acad Sci USA 92, 146–150.
Zhang, X., Li, T. 2007. Improved particle swarm optimization algorithm for 2D protein folding prediction. In: Proceedings of the 1st International Conference on Bioinformatics and Biomedical Engineering, China, 53–56.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mansour, N., Kanj, F. & Khachfe, H. Particle swarm optimization approach for protein structure prediction in the 3D HP model. Interdiscip Sci Comput Life Sci 4, 190–200 (2012). https://doi.org/10.1007/s12539-012-0131-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-012-0131-z